Disrupted uromodulin trafficking is rescued by targeting TMED cargo receptors
- PMID: 39680459
- PMCID: PMC11645142
- DOI: 10.1172/JCI180347
Disrupted uromodulin trafficking is rescued by targeting TMED cargo receptors
Abstract
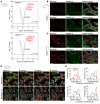
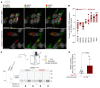
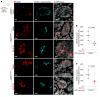
The trafficking dynamics of uromodulin (UMOD), the most abundant protein in human urine, play a critical role in the pathogenesis of kidney disease. Monoallelic mutations in the UMOD gene cause autosomal dominant tubulointerstitial kidney disease (ADTKD-UMOD), an incurable genetic disorder that leads to kidney failure. The disease is caused by the intracellular entrapment of mutant UMOD in kidney epithelial cells, but the precise mechanisms mediating disrupted UMOD trafficking remain elusive. Here, we report that transmembrane Emp24 protein transport domain-containing (TMED) cargo receptors TMED2, TMED9, and TMED10 bind UMOD and regulate its trafficking along the secretory pathway. Pharmacological targeting of TMEDs in cells, in human kidney organoids derived from patients with ADTKD-UMOD, and in mutant-UMOD-knockin mice reduced intracellular accumulation of mutant UMOD and restored trafficking and localization of UMOD to the apical plasma membrane. In vivo, the TMED-targeted small molecule also mitigated ER stress and markers of kidney damage and fibrosis. Our work reveals TMED-targeting small molecules as a promising therapeutic strategy for kidney proteinopathies.
Keywords: Genetic diseases; Nephrology; Protein misfolding; Protein traffic.
Conflict of interest statement
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

