Mortality-associated plasma proteome dynamics in a prospective multicentre sepsis cohort
- PMID: 39681038
- PMCID: PMC11714398
- DOI: 10.1016/j.ebiom.2024.105508
Mortality-associated plasma proteome dynamics in a prospective multicentre sepsis cohort
Abstract
Background: Sepsis remains a leading cause of mortality in intensive care units. Understanding the dynamics of the plasma proteome of patients with sepsis is critical for improving prognostic and therapeutic strategies.
Methods: This prospective, multicentre observational cohort study included 363 patients with sepsis recruited from five university hospitals in Germany between March 2018 and April 2023. Plasma samples were collected on days 1 and 4 after sepsis diagnosis, and proteome analysis was performed using mass spectrometry. Classical statistical methods and machine learning (random forest) were employed to identify proteins associated with 30-day survival outcomes.
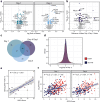
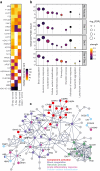
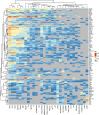
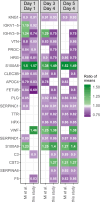
Findings: Out of 363 patients, 224 (62%) survived, and 139 (38%) did not survive the 30-day period. Proteomic analysis revealed significant differences in 87 proteins on day 1 and 95 proteins on day 4 between survivors and non-survivors. Additionally, 63 proteins were differentially regulated between day 1 and day 4 in the two groups. The identified protein networks were primarily related to blood coagulation, immune response, and complement activation. The random forest classifier achieved an area under the receiver operating characteristic curve of 0.75 for predicting 30-day survival. The results were compared and partially validated with an external sepsis cohort.
Interpretation: This study describes temporal changes in the plasma proteome associated with mortality in sepsis. These findings offer new insights into sepsis pathophysiology, emphasizing the innate immune system as an underexplored network, and may inform the development of targeted therapeutic strategies.
Funding: European Regional Development Fund of the European Union. The State of North Rhine-Westphalia, Germany.
Keywords: Feature importance; Machine learning; Mass spectrometry; Proteomics; Sepsis.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests AZ received payments for grants, lectures or consulting from Baxter BioMerieux, Bayer, AM Pharma, Novartis, RenalGuard, Alexion, Baxter, Paion and Viatris. JCK received funding from Danaher Beacon. AZ has leadership or fiduciary role in IARS, DIVI and DGAI. ME is employee of Ruhr-University Bochum.
Figures
References
-
- Raith E.P., Udy A.A., Bailey M., et al. Prognostic accuracy of the SOFA score, SIRS criteria, and qSOFA score for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA. 2017;317(3):290–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

