Development of a RIPK1 degrader to enhance antitumor immunity
- PMID: 39681571
- PMCID: PMC11649918
- DOI: 10.1038/s41467-024-55006-2
Development of a RIPK1 degrader to enhance antitumor immunity
Abstract
The scaffolding function of receptor interacting protein kinase 1 (RIPK1) confers intrinsic and extrinsic resistance to immune checkpoint blockades (ICBs) and emerges as a promising target for improving cancer immunotherapies. To address the challenge posed by a poorly defined binding pocket within the intermediate domain of RIPK1, here we harness proteolysis targeting chimera (PROTAC) technology to develop a RIPK1 degrader, LD4172. LD4172 exhibits potent and selective RIPK1 degradation both in vitro and in vivo. Degradation of RIPK1 by LD4172 triggers immunogenic cell death, enhances tumor-infiltrating lymphocyte responses, and sensitizes tumors to anti-PD1 therapy in female C57BL/6J mice. This work reports a RIPK1 degrader that serves as a chemical probe for investigating the scaffolding functions of RIPK1 and as a potential therapeutic agent to enhance tumor responses to ICBs therapy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J.W. is a co-founder of Chemical Biology Probes, LLC, and serves as a consultant for CoRegen Inc. X.Y., D.L., and J.W. are inventors on a patent covering RIPK1 degraders reported in this work, titled “Novel RIPK1 Kinase-Targeting PROTACs and Methods of Use Thereof”, with the identification number WO2022120118A1. The remaining authors declare no competing interests.
Figures
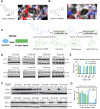

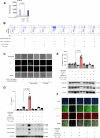

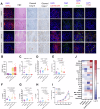
Update of
-
Development of a First-in-Class RIPK1 Degrader to Enhance Antitumor Immunity.bioRxiv [Preprint]. 2024 Mar 29:2024.03.25.586133. doi: 10.1101/2024.03.25.586133. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 16;15(1):10683. doi: 10.1038/s41467-024-55006-2. PMID: 38590362 Free PMC article. Updated. Preprint.
-
Development of a First-in-Class RIPK1 Degrader to Enhance Antitumor Immunity.Res Sq [Preprint]. 2024 Apr 8:rs.3.rs-4156736. doi: 10.21203/rs.3.rs-4156736/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Dec 16;15(1):10683. doi: 10.1038/s41467-024-55006-2. PMID: 38659866 Free PMC article. Updated. Preprint.
References
-
- Korman, A. J., Garrett-Thomson, S. C. & Lonberg, N. The foundations of immune checkpoint blockade and the ipilimumab approval decennial. Nat. Rev. Drug Discov.21, 509–528 (2022). - PubMed
-
- Vesely, M. D., Zhang, T. & Chen, L. Resistance mechanisms to anti-PD cancer immunotherapy. Annu. Rev. Immunol.40, 45–74 (2022). - PubMed
-
- Upadhaya, S., Neftelinov, S. T., Hodge, J. & Campbell, J. Challenges and opportunities in the PD1/PDL1 inhibitor clinical trial landscape. Nat. Rev. Drug Discov.21, 482–483 (2022). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R01-CA250503/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- RP220480/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- P30 CA125123/CA/NCI NIH HHS/United States
- R01 CA268518/CA/NCI NIH HHS/United States
- R01-CA268518/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Miscellaneous

