Differential characteristics of vaginal versus endometrial microbiota in IVF patients
- PMID: 39681607
- PMCID: PMC11649765
- DOI: 10.1038/s41598-024-82466-9
Differential characteristics of vaginal versus endometrial microbiota in IVF patients
Abstract
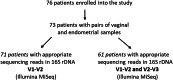
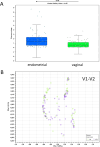
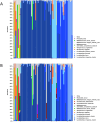
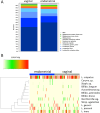
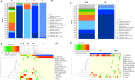
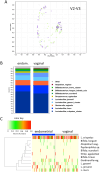
Abnormal female reproductive tract microbiota are associated with gynecological disorders such as endometriosis or chronic endometritis and may affect reproductive outcomes. However, the differential diagnostic utility of the vaginal or the endometrial microbiome and the impact of important technical covariates such as the choice of hypervariable regions for 16 S rRNA sequencing remain to be characterized. The aim of this retrospective study was to compare vaginal and endometrial microbiomes in IVF patients diagnosed with implantation failure (IF) and/or recurrent pregnancy loss (RPL) and to investigate the overlap between established vaginal and endometrial microbiome classification schemes. An additional aim was to characterize to which extent the choice of V1-V2 or V2-V3 16 S rRNA sequencing schemes influences the characterization of genital microbiomes. We compared microbiome composition based on V1-V2 rRNA sequencing between matched vaginal smear and endometrial pipelle-obtained biopsy samples (n = 71); in a sub-group (n = 61), we carried out a comparison between V1-V2 and V2-V3 rRNA sequencing. Vaginal and endometrial microbiomes were found to be Lactobacillus-dominated in the majority of patients, with the most abundant Lactobacillus species typically shared between sample types of same patient. Endometrial microbiomes were found to be more diverse than vaginal microbiomes (average Shannon entropy = 1.89 v/s 0.75, p = 10-5) and bacterial species such as Corynebacterium sp., Staphylococcus sp., Prevotella sp. and Propionibacterium sp. were found to be enriched in the endometrial samples. The use of two widely used clinical classification schemes to detect microbiome dysbiosis in the reproductive tract often led to inconsistent results vaginal community state type (CST) IV, which is associated with bacterial vaginosis, was detected in 9.8% of patients; however, 31,0% of study participants had a non-Lactobacillus-dominated (NLD) endometrial microbiome associated with unfavorable reproductive outcomes. Results based on V2-V3 rRNA sequencing were generally consistent with V1-V2-based; differences were observed for a small number of species, e.g. Bifidobacterium sp., Propionibacterium sp. and Staphylococcus sp. and with respect to slightly increased detection rates of CST IV and NLD. Our study showed that endometrial microbiomes differ substantially from their vaginal counterparts, the application of a trans-cervical sampling method notwithstanding. Characterization of endometrial microbiomes may contribute to the improved detection of women with an unfavorable reproductive outcome prognosis in IVF patients..
Keywords: Endometrial; Genital microbiota; IVF patients; RIF; RM; Vaginal.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

