TIGIT and PD-L1 co-blockade promotes clonal expansion of multipotent, non-exhausted antitumor T cells by facilitating co-stimulation
- PMID: 39681653
- PMCID: PMC11663793
- DOI: 10.1038/s43018-024-00870-6
TIGIT and PD-L1 co-blockade promotes clonal expansion of multipotent, non-exhausted antitumor T cells by facilitating co-stimulation
Erratum in
-
Publisher Correction: TIGIT and PD-L1 co-blockade promotes clonal expansion of multipotent, non-exhausted antitumor T cells by facilitating co-stimulation.Nat Cancer. 2025 Feb;6(2):405. doi: 10.1038/s43018-025-00908-3. Nat Cancer. 2025. PMID: 39825000 Free PMC article. No abstract available.
Abstract
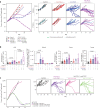
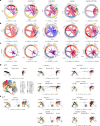
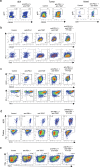
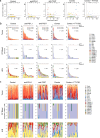
Blockade of immune checkpoints PD-1 and TIGIT has demonstrated activity in mouse tumor models and human patients with cancer. Although these coinhibitory receptors can restrict signaling in CD8+ T cells by regulating their associated co-stimulatory receptors CD28 and CD226, the functional consequences of combining PD-1 and TIGIT blockade remain poorly characterized. In mouse tumor models, we show that combination blockade elicited CD226-driven clonal expansion of tumor antigen-specific CD8+ T cells. The expanded clones emerged from a population of stem-like cells in draining lymph nodes, entering the blood as a previously unidentified single-phenotype, multiclonal population. Upon reaching the tumor, these transiting cells expanded further and differentiated into effector or exhausted T cells, with combination blockade restricting entry into the exhaustion pathway by favoring co-stimulation. Thus, PD-1 and TIGIT inhibition helps shape the repertoire of tumor-reactive CD8+ T cells in draining lymph nodes and determines their immunological fate in the tumor to enhance therapeutic benefit. Analysis of clinical trial samples suggests a similar mechanism may also occur in patients with cancer.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: all authors are employees of Genentech, a member of the Roche group, which develops and markets drugs for profit.
Figures













References
-
- Mellman, I., Chen, D. S., Powles, T. & Turley, S. J. The cancer-immunity cycle: indication, genotype, and immunotype. Immunity56, 2188–2205 (2023). - PubMed
-
- Cho, B. C. et al. Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol.23, 781–792 (2022). - PubMed
-
- Johnston, R. J. et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell26, 923–937 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

