qPCR-based quantification reveals high plant host-specificity of endophytic colonization levels in leaves
- PMID: 39682006
- PMCID: PMC11744438
- DOI: 10.1002/ajb2.16448
qPCR-based quantification reveals high plant host-specificity of endophytic colonization levels in leaves
Abstract
Premise: Despite the high functional importance of endophytes, we still have limited understanding of the biotic and abiotic factors that influence colonization of plant hosts along major ecological gradients and lack quantitative estimates of their colonization extent. In this study, we hypothesized that the developmental stage of the ecosystem will affect the levels of bacterial and fungal endophytic assemblages in the foliar endosphere.
Methods: We quantified levels of bacterial and fungal endophytes in leaves of four plant hosts at four stages of vegetation succession using an optimized qPCR protocol with bacteria-specific 16S and fungi-targeting primers.
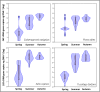
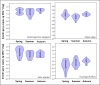
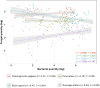
Results: (1) The ecosystem developmental stage did not have a significant effect on the colonization levels of bacterial or fungal endophytes. (2) Colonization levels by bacterial and fungal endophytes were governed by different mechanisms. (3) Endophytic colonization levels and their relationship to foliar tissue stoichiometry were highly host specific.
Conclusions: Quantifying colonization levels is important in the study of endophytic ecology, and the fast, relatively low-cost qPCR-based method can supply useful ecological information, which can significantly enhance the interpretation potential of descriptive data generated, for example, by next-generation sequencing.
Keywords: cell counts; ecological succession; foliar endophyte; fungi‐bacteria ratios; qPCR; soil chronosequence.
© 2024 The Author(s). American Journal of Botany published by Wiley Periodicals LLC on behalf of Botanical Society of America.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Plant Identity Influences Foliar Fungal Symbionts More Than Elevation in the Colorado Rocky Mountains.Microb Ecol. 2019 Oct;78(3):688-698. doi: 10.1007/s00248-019-01336-4. Epub 2019 Feb 4. Microb Ecol. 2019. PMID: 30715579
-
The effects of Pinus sylvestris L. geographical origin on the community and co-occurrence of fungal and bacterial endophytes in a common garden experiment.Microbiol Spectr. 2024 Oct 3;12(10):e0080724. doi: 10.1128/spectrum.00807-24. Epub 2024 Sep 9. Microbiol Spectr. 2024. PMID: 39248476 Free PMC article.
-
Plant growth promotion of Miscanthus × giganteus by endophytic bacteria and fungi on non-polluted and polluted soils.World J Microbiol Biotechnol. 2018 Mar 13;34(3):48. doi: 10.1007/s11274-018-2426-7. World J Microbiol Biotechnol. 2018. PMID: 29536268
-
Endophytic fungi: Unravelling plant-endophyte interaction and the multifaceted role of fungal endophytes in stress amelioration.Plant Physiol Biochem. 2024 Jan;206:108174. doi: 10.1016/j.plaphy.2023.108174. Epub 2023 Nov 17. Plant Physiol Biochem. 2024. PMID: 38070242 Review.
-
Fungal endophytes can modulate plant invasion.Biol Rev Camb Philos Soc. 2024 Oct;99(5):1652-1671. doi: 10.1111/brv.13085. Epub 2024 Apr 17. Biol Rev Camb Philos Soc. 2024. PMID: 38629189 Review.
Cited by
-
Genome-Driven Functional Validation of Bacillus amyloliquefaciens Strain MEPW12: A Multifunctional Endophyte for Sustainable Sweet Potato Cultivation.Microorganisms. 2025 Jun 6;13(6):1322. doi: 10.3390/microorganisms13061322. Microorganisms. 2025. PMID: 40572210 Free PMC article.
References
-
- Apigo, A. , and Oono R.. 2018. Dimensions of host specificity in foliar fungal endophytes. In: Pirttilä, A. , Frank, A. (eds) Endophytes of Forest Trees. Forestry Sciences, vol 86. Springer, Cham.
-
- Apigo, A. , and Oono R.. 2022. Plant abundance, but not plant evolutionary history, shapes patterns of host specificity in foliar fungal endophytes. Ecosphere 13: e03879.
-
- Bazzaz, F. A. 1979. The physiological ecology of plant succession. Annual Review of Ecology, Evolution, and Systematics 10: 351–371.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

