Plakin Expression in Serous Epithelial Ovarian Cancer Has the Potential to Impede Metastatic Spread and Epithelial-Mesenchymal Transition: A Comparative Expression Analysis of Immunohistochemical and In Silico Datasets
- PMID: 39682273
- PMCID: PMC11639978
- DOI: 10.3390/cancers16234087
Plakin Expression in Serous Epithelial Ovarian Cancer Has the Potential to Impede Metastatic Spread and Epithelial-Mesenchymal Transition: A Comparative Expression Analysis of Immunohistochemical and In Silico Datasets
Abstract
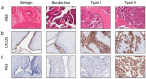
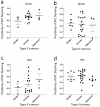
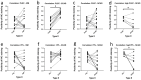
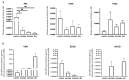
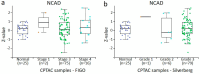
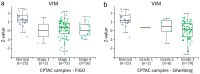
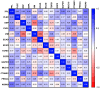
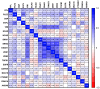
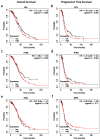
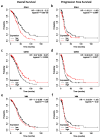
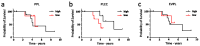
Epithelial ovarian cancer is aggressive and causes high mortality among women worldwide. Members of the plakin family are essential to maintain cytoskeletal integrity and key cellular processes. In this study we characterised the expression of plakins, particularly plectin (PLEC), periplakin (PPL), envoplakin (EVPL), and EMT-related proteins by immunohistochemistry in n = 48 patients' samples to evaluate a potential correlation of plakin expression with EMT as EOC progresses. These tissue plakin and EMT expression analyses were further evaluated by in vitro cell line expression and correlated with the expression of these molecules using publicly available datasets such as Cancer Genome Atlas (TCGA) and Clinical Proteome Tumour Analysis Consortium (CPTAC) datasets. We demonstrate that the expression of PPL and PLEC plakins is decreased in high-grade compared to low-grade EOCs with mixed EMT marker protein expression. This is supported by the correlation of high PPL and PLEC expression with an epithelial rather than mesenchymal phenotype. Our data suggest a partial loss of plakin expression as EOC tumours progress. This may impact the connections of plakins with membrane-bound receptors, which impede the downstream signalling required for the initiation of EMT as the tumours progress.
Keywords: ascites; epithelial ovarian cancer; epithelial–mesenchymal transition; in silico datasets; metastasis; plakins.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R.L., Soerjomataram I., Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–263. - PubMed
-
- Latifi A., Escalona R., Quinn M.A., Thompson E.W., Findlay J.K., Ahmed N. Distinct molecular signature of recurrent ovarian tumor cells isolated from the ascites of advanced-stage serous ovarian cancer patients. J. Cancer Stem Cell Res. 2014;2:e10061-20. doi: 10.14343/JCSCR.2014.2e1006. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

