Temporal Changes in Faecal Microbiota Composition and Diversity in Dairy Cows Supplemented with a Lactobacillus-Based Direct-Fed Microbial
- PMID: 39682401
- PMCID: PMC11640597
- DOI: 10.3390/ani14233437
Temporal Changes in Faecal Microbiota Composition and Diversity in Dairy Cows Supplemented with a Lactobacillus-Based Direct-Fed Microbial
Abstract
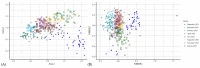
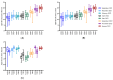
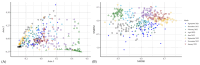
The rumen microbiota of dairy cows plays a crucial role in fermenting fibrous material, essential for nutrient extraction and overall productivity, detoxification of anti-nutritional toxic compounds, synthesis of vital nutrients, and is essential for optimal animal health. This study investigated the impact of Lentilactobacillus-, Lactocaseibacillus-, and Lacticaseibacillus-based direct-fed microbial (DFM) supplementation on dairy cows' faecal microbial composition and diversity. The study was carried out on a commercial dairy farm using 50 Holstein-Friesian cows randomly assigned into control (CON) and treatment (TRT) groups. Faecal samples were collected directly from the rectum every two months from September 2021 to January 2023. The bacterial 16S rRNA gene and fungal ITS-1 regions were amplified, sequenced, and analysed. Microbial diversity was assessed through alpha- and beta-diversity metrics. Linear discriminant analysis effect size (LEfSe) was performed to identify which taxa were driving the changes seen in the microbiota over time and treatment. Bacteroidaceae were the most prevalent bacterial family, followed by Lachnospiraceae and Muribaculaceae in both CON and TRT cows. Ascomycota, Basidiomycota, and Mucoromycota were the dominant three fungal phyla in the faeces of both CON and TRT cows. Bacterial genera Fructilactobacillus was abundant in the CON and Absicoccus in the TRT groups. Fungal taxa Chaetothryriales_incertae_sedis and Pseudomentella were absent in the faeces of TRT cows. Significant temporal and specific taxonomic differences were observed between the CON and TRT groups. The study's findings underscore the dynamic nature of microbial communities and the importance of targeted dietary interventions. Further research is necessary to elucidate these microbial shifts, long-term impacts, and functional implications, aiming to optimise ruminant nutrition and enhance dairy cow performance.
Keywords: DFM; dairy cows; faecal microbiota; microbial diversity; temporal changes.
Conflict of interest statement
M.S. and O.R.-G. were employed by Terragen Biotech Pty Ltd., which funded this study and assisted in the administrative component of this multiyear study. All remaining co-authors declare no conflicts of interest. The funders had no role in the design of the study, in the analyss or interpretation of the data, in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Samtiya M., Aluko R.E., Dhewa T. Plant food anti-nutritional factors and their reduction strategies: An overview. Food Prod. Process. Nutr. 2020;2:6. doi: 10.1186/s43014-020-0020-5. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

