Integron-Mediated Antimicrobial Resistance and Virulence Factors in Salmonella Typhimurium Isolated from Poultry
- PMID: 39682448
- PMCID: PMC11640015
- DOI: 10.3390/ani14233483
Integron-Mediated Antimicrobial Resistance and Virulence Factors in Salmonella Typhimurium Isolated from Poultry
Abstract
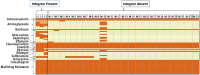
This study investigates antimicrobial-resistant (AMR) Salmonella Typhimurium in poultry, focusing on how class I integrons contribute to AMR and virulence. Using whole genome sequencing, researchers analyzed 26 S. Typhimurium isolates from U.S. poultry, finding that three isolates contained integrons (1000 base pairs each). These integron-positive isolates exhibited significantly higher resistance to beta-lactams, phenicols, and tetracyclines compared to integron-free isolates (p = 0.004, 0.009, and 0.02, respectively) and harbored genes like ges, imp, and oxa, which are linked to extended-spectrum beta-lactamase resistance. Most AMR gene classes (64%) were chromosome-based, with integron-positive isolates showing a broader array of resistance genes, including catB and tetA. Integron-bearing isolates had higher occurrences of bacteriocin genes and specific AMR genes like aminoglycoside and beta-lactam resistance genes, while integron-free isolates had more fimbrial and pilus genes. The presence of integrons may trend with increased AMR genes and virulence factors, highlighting the role of integron screening in enhancing AMR surveillance and reducing the need for high-priority antimicrobial treatments in poultry. These findings could support better AMR stewardship practices in poultry production, potentially lowering infection risks in humans and livestock.
Keywords: Salmonella Typhimurium; antimicrobial resistance; integrons; poultry; virulence factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. World Health Organization; Geneva, Switzerland: 2024.
Grants and funding
LinkOut - more resources
Full Text Sources

