Overexpression of Egr1 Transcription Regulator Contributes to Schwann Cell Differentiation Defects in Neural Crest-Specific Adar1 Knockout Mice
- PMID: 39682701
- PMCID: PMC11639873
- DOI: 10.3390/cells13231952
Overexpression of Egr1 Transcription Regulator Contributes to Schwann Cell Differentiation Defects in Neural Crest-Specific Adar1 Knockout Mice
Abstract
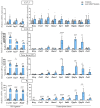
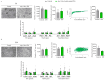
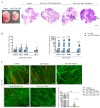
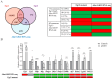
Adenosine deaminase acting on RNA 1 (ADAR1) is the principal enzyme for the adenosine-to-inosine RNA editing that prevents the aberrant activation of cytosolic nucleic acid sensors by endogenous double stranded RNAs and the activation of interferon-stimulated genes. In mice, the conditional neural crest deletion of Adar1 reduces the survival of melanocytes and alters the differentiation of Schwann cells that fail to myelinate nerve fibers in the peripheral nervous system. These myelination defects are partially rescued upon the concomitant removal of the Mda5 antiviral dsRNA sensor in vitro, suggesting implication of the Mda5/Mavs pathway and downstream effectors in the genesis of Adar1 mutant phenotypes. By analyzing RNA-Seq data from the sciatic nerves of mouse pups after conditional neural crest deletion of Adar1 (Adar1cKO), we here identified the transcription factors deregulated in Adar1cKO mutants compared to the controls. Through Adar1;Mavs and Adar1cKO;Egr1 double-mutant mouse rescue analyses, we then highlighted that the aberrant activation of the Mavs adapter protein and overexpression of the early growth response 1 (EGR1) transcription factor contribute to the Adar1 deletion associated defects in Schwann cell development in vivo. In silico and in vitro gene regulation studies additionally suggested that EGR1 might mediate this inhibitory effect through the aberrant regulation of EGR2-regulated myelin genes. We thus demonstrate the role of the Mda5/Mavs pathway, but also that of the Schwann cell transcription factors in Adar1-associated peripheral myelination defects.
Keywords: ADAR1; EGR1; MAVS; Schwann cells; differentiation; neural crest.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

