Gene-Silencing Therapeutic Approaches Targeting PI3K/Akt/mTOR Signaling in Degenerative Intervertebral Disk Cells: An In Vitro Comparative Study Between RNA Interference and CRISPR-Cas9
- PMID: 39682777
- PMCID: PMC11640589
- DOI: 10.3390/cells13232030
Gene-Silencing Therapeutic Approaches Targeting PI3K/Akt/mTOR Signaling in Degenerative Intervertebral Disk Cells: An In Vitro Comparative Study Between RNA Interference and CRISPR-Cas9
Abstract
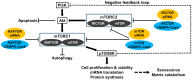
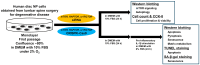
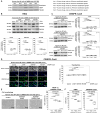
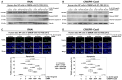
The mammalian target of rapamycin (mTOR), a serine/threonine kinase, promotes cell growth and inhibits autophagy. The following two complexes contain mTOR: mTORC1 with the regulatory associated protein of mTOR (RAPTOR) and mTORC2 with the rapamycin-insensitive companion of mTOR (RICTOR). The phosphatidylinositol 3-kinase (PI3K)/Akt/mTOR signaling pathway is important in the intervertebral disk, which is the largest avascular, hypoxic, low-nutrient organ in the body. To examine gene-silencing therapeutic approaches targeting PI3K/Akt/mTOR signaling in degenerative disk cells, an in vitro comparative study was designed between small interfering RNA (siRNA)-mediated RNA interference (RNAi) and clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated protein 9 (Cas9) gene editing. Surgically obtained human disk nucleus pulposus cells were transfected with a siRNA or CRISPR-Cas9 plasmid targeting mTOR, RAPTOR, or RICTOR. Both of the approaches specifically suppressed target protein expression; however, the 24-h transfection efficiency differed by 53.8-60.3% for RNAi and 88.1-89.3% for CRISPR-Cas9 (p < 0.0001). Targeting mTOR, RAPTOR, and RICTOR all induced autophagy and inhibited apoptosis, senescence, pyroptosis, and matrix catabolism, with the most prominent effects observed with RAPTOR CRISPR-Cas9. In the time-course analysis, the 168-h suppression ratio of RAPTOR protein expression was 83.2% by CRISPR-Cas9 but only 8.8% by RNAi. While RNAi facilitates transient gene knockdown, CRISPR-Cas9 provides extensive gene knockout. Our findings suggest that RAPTOR/mTORC1 is a potential therapeutic target for degenerative disk disease.
Keywords: RNA interference (RNAi); autophagy; clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein 9 (Cas9); disk degeneration; gene-silencing therapy; intervertebral disk; nucleus pulposus (NP) cells; phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling; small interfering RNA (siRNA); spine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




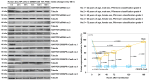
References
-
- Livshits G., Popham M., Malkin I., Sambrook P.N., Macgregor A.J., Spector T., Williams F.M. Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: The UK Twin Spine Study. Ann. Rheum. Dis. 2011;70:1740–1745. doi: 10.1136/ard.2010.137836. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

