Genome-Wide Identification, Classification, Expression Analysis, and Screening of Drought and Heat Resistance-Related Candidates of the Rboh Gene Family in Wheat
- PMID: 39683170
- PMCID: PMC11644269
- DOI: 10.3390/plants13233377
Genome-Wide Identification, Classification, Expression Analysis, and Screening of Drought and Heat Resistance-Related Candidates of the Rboh Gene Family in Wheat
Abstract
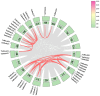
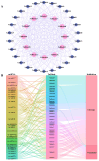
Plant respiratory burst oxidase homologs (Rbohs) are key enzymes that produce reactive oxygen species (ROS), which serve as signaling molecules regulating plant growth and stress responses. In this study, 39 TaRboh genes (TaRboh01-TaRboh39) were identified. These genes were distributed unevenly among the wheat genome's fourteen chromosomes, with the exception of homoeologous group 2 and 7 and chromosomes 4A, as well as one unidentified linkage group (Un). TaRbohs were classified into ten distinct clades, each sharing similar motif compositions and gene structures. The promoter regions of TaRbohs contained cis-elements related to hormones, growth and development, and stresses. Furthermore, five TaRboh genes (TaRboh26, TaRboh27, TaRboh31, TaRboh32, and TaRboh34) exhibited strong evolutionary conservation. Additionally, a Ka/Ks analysis confirmed that purifying selection was the predominant force driving the evolution of these genes. Expression profiling and qPCR results further indicated differential expression patterns of TaRboh genes between heat and drought stresses. TaRboh11, TaRboh20, TaRboh22, TaRboh24, TaRboh29, and TaRboh34 were significantly upregulated under multiple stress conditions, whereas TaRboh30 was only elevated in response to drought stress. Collectively, our findings provide a systematic analysis of the wheat Rboh gene family and establish a theoretical framework for our future research on the role of Rboh genes in response to heat and drought stress.
Keywords: Rbohs; Triticum aestivum; abiotic stress; expression patterns; reactive oxygen species.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Yu S., Kakar K.U., Yang Z., Nawaz Z., Lin S., Guo Y., Ren X.-L., Baloch A.A., Han D. Systematic Study of the Stress-Responsive Rboh Gene Family in Nicotiana Tabacum: Genome-Wide Identification, Evolution and Role in Disease Resistance. Genomics. 2020;112:1404–1418. doi: 10.1016/j.ygeno.2019.08.010. - DOI - PubMed
Grants and funding
- 32372124, 32300456, 82304652/National Natural Science Foundation of China
- 2022M721101, 2021M701160, 2023M731065/China Postdoctoral Science Foundation
- 2023JJ40132, 2022JJ40051, 2023JJ40199/Hunan Provincial Department of Science and Technology
- kq2202149/Changsha Natural Science Foundation
- CSTB2023NSCQ-MSX0542, CSTB2023NSCQ-MSX1031, CSTB2022NSCQ-MSX0517, CSTB2022NSCQ-MSX1138/Natural Science Foundation of Chongqing, China
LinkOut - more resources
Full Text Sources

