A Bifunctional Nuclease Promotes the Infection of Zucchini Yellow Mosaic Virus in Watermelon by Targeting P3
- PMID: 39683224
- PMCID: PMC11644367
- DOI: 10.3390/plants13233431
A Bifunctional Nuclease Promotes the Infection of Zucchini Yellow Mosaic Virus in Watermelon by Targeting P3
Abstract
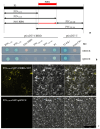
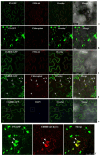
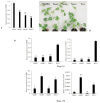
Potyviral P3 is involved in viral replication, movement, and pathogenicity; however, its biochemical function is unknown. In this study, the P3 of the zucchini yellow mosaic virus (ZYMV) interacted with ClBBD, a protein with high ortholog bifunctional nuclease activity, in watermelon. The binding site was shown via yeast two-hybrid screening and BiFC assay to be located at the N-terminus of P3 rather than P3N-PIPO. ClBBD localized predominantly to the chloroplast and plasma membrane. ZYMV P3 was also present in the nucleus and cytoplasm as aggregates. When co-expressed with P3 in tobacco, ClBBD formed aggregates with P3 in the cytoplasm. The knockdown of ClBBD using the VIGS vector pV190 and challenge with ZYMV revealed a positive correlation between viral accumulation and ClBBD expression, indicating that ClBBD reduces the resistance of watermelon to ZYMV. Furtherly, we found that when P3 and ClBBD were transiently co-expressed in tobacco, the level of P3 was significantly higher than that when it was expressed alone or co-expressed with GUS. It inferred that ClBBD may be able to stabilize the expression of P3. Overall, the results suggest that the interaction of P3 with ClBBD promotes virus infection, and ClBBD may be involved in stabilizing the expression level of P3.
Keywords: P3; bifunctional nuclease; interaction; watermelon; zucchini yellow mosaic virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Desbiez C., Lecoq H. Zucchini yellow mosaic virus. Plant Pathol. 1997;46:809–829. doi: 10.1046/j.1365-3059.1997.d01-87.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

