M2BPgs-HCC: An Automated Multilectin Bead Array Indicating Aberrant Glycosylation Signatures Toward Hepatitis C Virus-Associated Hepatocellular Carcinoma Prognosis
- PMID: 39683799
- PMCID: PMC11643838
- DOI: 10.3390/molecules29235640
M2BPgs-HCC: An Automated Multilectin Bead Array Indicating Aberrant Glycosylation Signatures Toward Hepatitis C Virus-Associated Hepatocellular Carcinoma Prognosis
Abstract
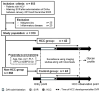
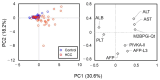
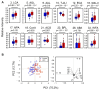
Regular monitoring of patients with a history of hepatitis C virus (HCV) infection is critical for the detection and management of hepatocellular carcinoma (HCC). Mac-2 binding protein glycosylation isomer (M2BPGi) has been used to monitor fibrosis progression and predict HCC. However, HCC prediction based on M2BPGi has not been optimized. Here, we identified HCC risk-related glycan signatures of M2BP using a newly developed automated bead array with multiplexed lectins. Among 955 patients with HCV who achieved sustained virological response following direct-acting antiviral treatment, we compared M2BP glycosylation from sera of 42 patients diagnosed with HCC during follow-up and 43 without HCC (control) by the lectin microarray. At the HCC observation point, we found significant differences in 17 lectins. Using an automated bead array with 12 of 17 lectins, a principal component analysis (PCA) biplot differentiated HCC from control, along the PC1 axis, explaining 75.2% of variance. Based on PC1, we generated a scoring formula for an HCC-related glycosylation signature on M2BP (M2BPgs-HCC), showing good diagnostic performance for HCC (p = 2.92 × 10-8, AUC = 0.829). This automated multilectin bead array improved the ability of M2BP to detect HCC, providing a candidate test for HCC surveillance in combination with other HCC markers.
Keywords: M2BPGi; glycosylation; hepatocellular carcinoma; lectin array.
Conflict of interest statement
The authors declare the following financial interests/personal relationships that may be considered potential competing interests: H.S. and T.O. have relationships with Precision System Science Co., Ltd.; M.M. received a lecture fee from Sysmex Co.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

