Enhancing Crop Resilience: The Role of Plant Genetics, Transcription Factors, and Next-Generation Sequencing in Addressing Salt Stress
- PMID: 39684248
- PMCID: PMC11640768
- DOI: 10.3390/ijms252312537
Enhancing Crop Resilience: The Role of Plant Genetics, Transcription Factors, and Next-Generation Sequencing in Addressing Salt Stress
Abstract
Salt stress is a major abiotic stressor that limits plant growth, development, and agricultural productivity, especially in regions with high soil salinity. With the increasing salinization of soils due to climate change, developing salt-tolerant crops has become essential for ensuring food security. This review consolidates recent advances in plant genetics, transcription factors (TFs), and next-generation sequencing (NGS) technologies that are pivotal for enhancing salt stress tolerance in crops. It highlights critical genes involved in ion homeostasis, osmotic adjustment, and stress signaling pathways, which contribute to plant resilience under saline conditions. Additionally, specific TF families, such as DREB, NAC (NAM, ATAF, and CUC), and WRKY, are explored for their roles in activating salt-responsive gene networks. By leveraging NGS technologies-including genome-wide association studies (GWASs) and RNA sequencing (RNA-seq)-this review provides insights into the complex genetic basis of salt tolerance, identifying novel genes and regulatory networks that underpin adaptive responses. Emphasizing the integration of genetic tools, TF research, and NGS, this review presents a comprehensive framework for accelerating the development of salt-tolerant crops, contributing to sustainable agriculture in saline-prone areas.
Keywords: crop improvement; genetic transcription factors (TFs); next-generation sequencing (NGS); salt stress; salt tolerance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
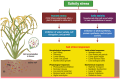
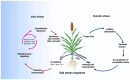
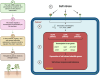
References
-
- FAO . Trends and Challenges. FAO; Rome, Italy: 2017. Agriculture organization of the United Nations the future of food and agriculture.
-
- Al-Khayri J.M., Ansari M.I., Singh A.K. Nanobiotechnology: Mitigation of Abiotic Stress in Plants. 1st ed. Springer; Cham, Switzerland: 2021. pp. 1–593.
-
- Singh A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2022;38:39–67. doi: 10.1111/sum.12772. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

