Unravelling the Signature Follicular Fluid Metabolites in Dairy Cattle Follicles Growing Under Negative Energy Balance: An In Vitro Approach
- PMID: 39684341
- PMCID: PMC11641226
- DOI: 10.3390/ijms252312629
Unravelling the Signature Follicular Fluid Metabolites in Dairy Cattle Follicles Growing Under Negative Energy Balance: An In Vitro Approach
Abstract
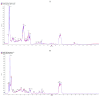
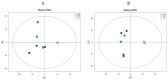
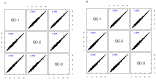
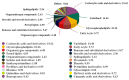
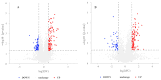
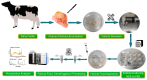
The astringent selection criteria for milk-oriented traits in dairy cattle have rendered these animals prone to various metabolic disorders. Postpartum lactational peak and reduced feed intake lead to negative energy balance in cattle. As a compensatory mechanism, cattle start mobilizing fat reserves to meet the energy demand for vital body functions. Consequently, diminished glucose concentrations and elevated ketone body levels lead to poor ovarian function. The impaired follicular development and subpar oocyte quality diminish the conception rates, which poses significant economic repercussions. Follicular fluid is integral to the processes of follicular growth and oocyte development. Hence, the present study was performed to identify potential alterations in metabolites in the follicular fluid under in vitro culture conditions mimicking negative energy balance. Our results revealed nine distinct metabolites exhibiting differential expression in follicular fluid under negative energy balance. The differentially expressed metabolites were predominantly associated with pathways related to amino acid metabolism, lipid metabolism, signal transduction mechanisms, and membrane transport, alongside other biological processes. The identified signature metabolites may be further validated to determine oocyte fitness subjected to in vitro fertilization or embryo production from slaughterhouse source ovaries.
Keywords: dairy cattle; follicular fluid; metabolites; negative energy balance.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- USDA Development of the Average Annual Milk Production Per Cow in the United States Since 1924. [(accessed on 19 September 2024)]; Available online: https://quickstats.nass.usda.gov/
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

