Single-Cell Analysis: A Method for In-Depth Phenotyping of Cells Involved in Asthma
- PMID: 39684345
- PMCID: PMC11641648
- DOI: 10.3390/ijms252312633
Single-Cell Analysis: A Method for In-Depth Phenotyping of Cells Involved in Asthma
Abstract
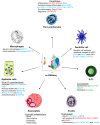
Asthma is a chronic inflammatory lung disease with high prevalence, making it one of the most common chronic conditions worldwide. Its pathophysiology is influenced by a range of genetic and environmental factors, resulting in a complex and heterogeneous disease profile. Asthma is primarily associated with a type 2 (T2) immune response, though non-T2 endotypes also contribute to disease pathology. Generally, asthma is characterized by the infiltration and activation of various cell types, including dendritic cells, eosinophils, innate lymphoid cells, lymphocytes, mast cells, and neutrophils, which participate in T1, T2, and T17 immune responses. Despite advances in understanding, many questions remain unresolved. Therefore, emerging omic techniques, such as single-cell RNA sequencing (scRNA-seq), offer novel insights into the underlying mechanisms of asthma and the roles of these immune cells. Recent scRNA-seq studies in asthma have identified multiple novel immune cell subtypes and clusters, suggesting their potential functions in disease pathology. The rapid advancement of scRNA-seq technology now enables in-depth investigation of individual cells within tissues, allowing for precise cell-type classification and detailed molecular profiling. Nonetheless, certain limitations persist, which require further refinement in future studies.
Keywords: allergic diseases; asthma; immune cells; single-cell RNA-seq.
Conflict of interest statement
VdP reports getting honoraria (advisory board, speaker) and/or institutional grant/research support from AstraZe©neca and GSK and holding unpaid Leadership or fiduciary role in committee in EAACI. The rest of authors declare no conflicts of interest.
Figures

References
-
- Lotvall J., Akdis C.A., Bacharier L.B., Bjermer L., Casale T.B., Custovic A., Lemanske R.F., Jr., Wardlaw A.J., Wenzel S.E., Greenberger P.A. Asthma endotypes: A new approach to classification of disease entities within the asthma syndrome. J. Allergy Clin. Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

