High- and Moderate-Risk Variants Among Breast Cancer Patients and Healthy Donors Enrolled in Multigene Panel Testing in a Population of Central Russia
- PMID: 39684352
- PMCID: PMC11641773
- DOI: 10.3390/ijms252312640
High- and Moderate-Risk Variants Among Breast Cancer Patients and Healthy Donors Enrolled in Multigene Panel Testing in a Population of Central Russia
Abstract
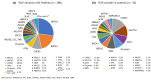
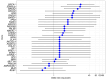
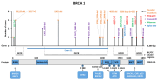
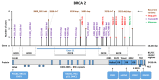
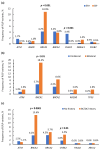
Assessments of breast cancer (BC) risk in carriers of pathogenic variants identified by gene panel testing in different populations are highly in demand worldwide. We performed target sequencing of 78 genes involved in DNA repair in 860 females with BC and 520 age- and family history-matched controls from Central Russia. Among BC patients, 562/860 (65.3%) were aged 50 years or less at the time of diagnosis. In total, 190/860 (22%) BC patients were carriers of 198 pathogenic/likely pathogenic (P/LP) variants in 30 genes, while among controls, 32/520 (6.2%) carriers of P/LP variants in 17 genes were identified. The odds ratio [95% confidence interval] was 16.3 [4.0-66.7] for BRCA1; 12.0 [2.9-45.9] for BRCA2; and 7.3 [0.9-56.7] for ATM (p < 0.05). Previously undescribed BRCA1/2, ATM, and PALB2 variants, as well as novel recurrent mutations, were identified. The contribution to BC susceptibility of truncating variants in the genes BARD1, RAD50, RAD51C, NBEAL1 (p. E1155*), and XRCC2 (p. P32fs) was evaluated. The BLM, NBN, and MUTYH genes did not demonstrate associations with BC risk. Finding deleterious mutations in BC patients is important for diagnosis and management; in controls, it opens up the possibility of prevention and early diagnostics.
Keywords: ATM; BRCA1/2; breast cancer risk; healthy controls; multigene panel testing; pathogenic variant; patients.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Bergstrom C., Pence C., Berg J., Partain N., Sadeghi N., Mauer C., Pirzadeh-Miller S., Gao A., Li H., Unni N., et al. Clinicopathological Features and Outcomes in Individuals with Breast Cancer and ATM, CHEK2, or PALB2 Mutations. Ann. Surg. Oncol. 2021;28:3383–3393. doi: 10.1245/s10434-020-09158-2. - DOI - PubMed
-
- Yadav S., Boddicker N.J., Na J., Polley E.C., Hu C., Hart S.N., Gnanaolivu R.D., Larson N., Holtegaard S., Huang H., et al. Contralateral Breast Cancer Risk Among Carriers of Germline Pathogenic Variants in ATM, BRCA1, BRCA2, CHEK2, and PALB2. J. Clin. Oncol. 2023;41:1703–1713. doi: 10.1200/JCO.22.01239. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

