Binding of MAP3773c Protein of Mycobacterium avium subsp. paratuberculosis in the Mouse Ferroportin1 Coding Region
- PMID: 39684399
- PMCID: PMC11641199
- DOI: 10.3390/ijms252312687
Binding of MAP3773c Protein of Mycobacterium avium subsp. paratuberculosis in the Mouse Ferroportin1 Coding Region
Abstract
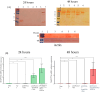
Mycobacterium avium subsp. paratuberculosis (MAP) is known to cause paratuberculosis. One notable protein, MAP3773c, plays a critical role in iron metabolism as a transcription factor. This study aims to investigate the binding affinity of MAP3773c to the chromatin of the Ferroportin1 (FPN1) gene in murine macrophage J774 A.1. We conducted a sequence alignment to identify potential interaction sites for MAP3773c. Following this, we used in silico analysis to predict binding interactions, complemented by electrophoretic mobility shift assay (EMSA) to confirm in vitro binding of MAP3773c. The map3773c gene was cloned into the pcDNA3.1 vector, with subsequent expression analysis carried out via Western blotting and real-time PCR. Chromatin immunoprecipitation (CHiP) assays were performed on transfected macrophages to confirm binding in the native chromatin context. Our in silico and in vitro analysis indicated that MAP3773c interacts with two binding motifs within the FPN1 coding region. The ChiP results provided additional validation, demonstrating the binding of MAP3773c to the FPN1 chromatin through successful amplification of the associated chromatin fragment via PCR. Our study demonstrated that MAP3773c binds to FPN1 and provides insight into the role of MAP3773c and its effect on host iron transport.
Keywords: Ferroportin1; MAP3773c; transcription factor; transcription regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

