USP8 Mutations Associated with Cushing's Disease Alter Protein Structure Dynamics
- PMID: 39684405
- PMCID: PMC11641493
- DOI: 10.3390/ijms252312697
USP8 Mutations Associated with Cushing's Disease Alter Protein Structure Dynamics
Abstract
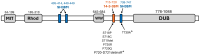
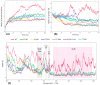
The adenomas in Cushing's disease frequently exhibit mutations in exon 14, within a binding motif for the regulatory protein 14-3-3 located between the catalytic domain (DUB), responsible for ubiquitin hydrolysis, and the WW-like domain that mediates autoinhibition, resulting in constantly active USP8. The exact molecular mechanism of deubiquitinase activity disruption in Cushing's disease remains unclear. To address this, Sanger sequencing of USP8 was performed to identify mutations in corticotropinomas. These mutations were subjected to computational screening, followed by molecular dynamics simulations to assess the structural alterations that might change the biological activity of USP8. Eight different variants of the USP8 gene were identified both within and outside the "hotspot" region. Six of these had previously been reported in Cushing's disease, while two were detected for the first time in our patients with CD. One of the two new variants, initially classified as benign during screening, was found in the neighboring SH3 binding motif at a distance of 20 amino acids. This variant demonstrated pathogenicity patterns similar to those of known pathogenic variants. All USP8 variants identified in our patients caused conformational changes in the USP8 protein in a similar manner. The identified mutations, despite differences in annotation results-including evolutionary conservation assessments, automated predictor data, and variations in localization within exon 14-exhibit similar patterns of protein conformational change. This suggests a pathogenic effect that contributes to the development of CD.
Keywords: 14-3-3 protein; Cushing’s disease; USP8 mutations; USP8 protein; docking; molecular dynamics; pituitary adenoma; protein modeling.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Lindholm J., Juul S., Jørgensen J.O.L., Astrup J., Bjerre P., Feldt-Rasmussen U., Hagen C., Jørgensen J., Kosteljanetz M., Kristensen L.Ø., et al. Incidence and Late Prognosis of Cushing’s Syndrome: A Population-Based Study. J. Clin. Endocrinol. Metab. 2001;86:117–123. doi: 10.1210/jc.86.1.117. - DOI - PubMed
-
- Gkourogianni A., Sinaii N., Jackson S.H., Karageorgiadis A.S., Lyssikatos C., Belyavskaya E., Keil M.F., Zilbermint M., Chittiboina P., Stratakis C.A., et al. Pediatric Cushing Disease: Disparities in Disease Severity and Outcomes in the Hispanic and African-American Populations. Pediatr. Res. 2017;82:272–277. doi: 10.1038/pr.2017.58. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

