Ascorbic Acid Ameliorates Molecular and Developmental Defects in Human-Induced Pluripotent Stem Cell and Cerebral Organoid Models of Fragile X Syndrome
- PMID: 39684429
- PMCID: PMC11641479
- DOI: 10.3390/ijms252312718
Ascorbic Acid Ameliorates Molecular and Developmental Defects in Human-Induced Pluripotent Stem Cell and Cerebral Organoid Models of Fragile X Syndrome
Abstract
Fragile X Syndrome (FX) is the most common form of inherited cognitive impairment and falls under the broader category of Autism Spectrum Disorders (ASD). FX is caused by a CGG trinucleotide repeat expansion in the non-coding region of the X-linked Fragile X Messenger Ribonucleoprotein 1 (FMR1) gene, leading to its hypermethylation and epigenetic silencing. Animal models of FX rely on the deletion of the Fmr1 gene, which fails to replicate the epigenetic silencing mechanism of the FMR1 gene observed in human patients. Human stem cells carrying FX repeat expansions have provided a better understanding of the basis of epigenetic silencing of FMR1. Previous studies have found that 5-Azacytidine (5Azac) can reverse this methylation; however, 5Azac can be toxic, which may limit its therapeutic potential. Here, we show that the dietary factor Ascorbic Acid (AsA) can reduce DNA methylation in the FMR1 locus and lead to an increase in FMR1 gene expression in FX iPSCs and cerebral organoids. In addition, AsA treatment rescued neuronal gene expression and morphological defects observed in FX iPSC-derived cerebral organoids. Hence, we demonstrate that the dietary co-factor AsA can partially revert the molecular and morphological defects seen in human FX models in vitro. Our findings have implications for the development of novel therapies for FX in the future.
Keywords: Ascorbic Acid; Autism Spectrum Disorders (ASD); FMR1; Fragile X Syndrome; cerebral organoids; gene silencing; induced pluripotent stem cells; methylation; neurodevelopmental disorders.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
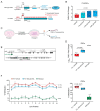
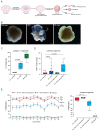
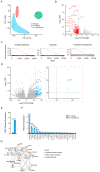
References
-
- Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-H. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

