Molecular Characteristics of Circ_002156 and Its Effects on Proliferation and Differentiation of Caprine Skeletal Muscle Satellite Cells
- PMID: 39684452
- PMCID: PMC11641368
- DOI: 10.3390/ijms252312745
Molecular Characteristics of Circ_002156 and Its Effects on Proliferation and Differentiation of Caprine Skeletal Muscle Satellite Cells
Abstract
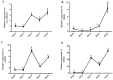
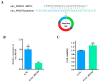
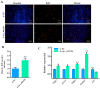
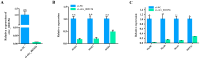
The proliferation and differentiation of skeletal muscle satellite cells (SMSCs) are responsible for the development of skeletal muscle. In our previous study, circ_002156 was found to be highly expressed in caprine Longissimus Dorsi muscle, but the regulatory role of the circular RNAs (circRNA) in goat SMSCs remains unclear. In this study, the authenticity of circ_002156 was validated, and its structurally characteristic and cellular localization as well as tissue expression of circ_002156 and its parent genes were investigated. Moreover, the effects of circ_002156 on the viability, proliferation, and differentiation of SMSCs were also studied. The circ_002156 is located on caprine chromosome 19 with a length of 36,478. The circRNA structurally originates from myosin heavy chain 2 (MYH2), MYH1, and MYH4 as well as intergenic sequences among the parent genes. RT-PCR and Sanger sequencing confirmed the authenticity of circ_002156. Most circ_002156 (55.5%) was expressed in the nuclei of SMSCs, while 44.5% of circ_002156 was located in the cytoplasm. The circ_002156 and its three parent genes had higher expression levels in the triceps brachii, quadriceps femoris, and longissimus dorsi muscle tissues than in the other five tissues. The expression of circ_002156 and its parent genes MYH1 and MYH4 reached the maximum on day 8 of differentiation, while MYH2 in expression reached the peak on day 4 after differentiation. The Pearson correlation coefficients revealed that circ_002156 had moderate or high positive correlations with the three parent genes in the expression of both quadriceps femoris muscle and SMSCs during different differentiation stages. The small interfering RNA circ_002156 (named si-circ_002156) remarkably increased the viability of the SMSCs. The si-circ_002156 also increased the number and parentage of Edu-labeled positive SMSCs as well as the expression levels of four cell proliferation marker genes. These suggest that circ_002156 inhibited the proliferation of SMSCs. Meanwhile, si-circ_002156 decreased the area of MyHC-labeled positive myotubes, the myotube fusion index, and myotube size as well as the expression of its three parent genes and four cell differentiation marker genes, suggesting a positive effect of circ_002156 on the differentiation of SMSCs. This study contributes to a better understanding of the roles of circ_002156 in the proliferation and differentiation of SMSCs.
Keywords: circ_002156; differentiation; goat; proliferation; skeletal muscle satellite cells.
Conflict of interest statement
We certify that there are no conflicts of interest with any financial organization regarding the material discussed in the manuscript.
Figures






References
MeSH terms
Substances
Grants and funding
- 22JR5RA829/the fund for Basic Research Creative Groups of Gansu Province
- GAU-XKTD-2022-21/the Discipline Team Project of Gansu Agricultural University
- Gaufx-02Y02/Fuxi Young Talents Fund of Gansu Agricultural University
- 2022-2-91/the Science and Technology project of Lanzhou city
- GAU2022-008/Gansu Agricultural University Postgraduate Joint-education Base Project
LinkOut - more resources
Full Text Sources

