Comprehensive Genome-Wide Identification and Expression Profiling of bHLH Transcription Factors in Areca catechu Under Abiotic Stress
- PMID: 39684646
- PMCID: PMC11640848
- DOI: 10.3390/ijms252312936
Comprehensive Genome-Wide Identification and Expression Profiling of bHLH Transcription Factors in Areca catechu Under Abiotic Stress
Abstract
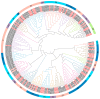
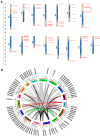
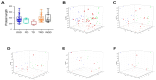
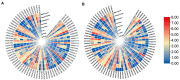
The basic helix-loop-helix (bHLH) transcription factor (TF) family, the second-largest among eukaryotes, is known for its evolutionary and functional diversity across plant species. However, bHLH genes have not yet been characterized in Areca catechu. In this study, we identified 76 AcbHLH genes, which exhibit a variety of physicochemical properties. Phylogenetic analysis revealed evolutionary relationships between Arabidopsis thaliana bHLH genes (AtbHLH) and their counterparts in A. catechu (AcbHLH). These analyses also highlighted conserved amino acid motifs (S, R, K, P, L, A, G, and D), conserved domains, and evolutionary changes, such as insertions, deletions, and exon gains or losses. Promoter analysis of AcbHLH genes revealed 76 cis-elements related to growth, phytohormones, light, and stress. Gene duplication analysis revealed four tandem duplications and twenty-three segmental duplications, while AcbHLH63 in the Areca genome exhibited significant synteny with bHLH genes from A. thaliana, Vitis vinifera, Solanum lycopersicum, Brachypodium distachyon, Oryza sativa, and Zea mays. Furthermore, relative expression analysis showed that under drought stress (DS), AcbHLH22, AcbHLH39, AcbHLH45, and AcbHLH58 showed distinct upregulation in leaves at specific time points, while all nine AcbHLH genes were upregulated in roots. Under salt stress (SS), AcbHLH22, AcbHLH39, AcbHLH45, and AcbHLH58 were upregulated in leaves, and AcbHLH22, AcbHLH34, and AcbHLH39 exhibited differential expression in roots at various time points. This study provides valuable insights into the bHLH superfamily in A. catechu, offering a solid foundation for further investigation into its role in responding to abiotic stresses.
Keywords: Areca catechu; abiotic stresses; bHLH gene family; expression pattern; genome-wide analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Lai X., Vega-Léon R., Hugouvieux V., Blanc-Mathieu R., van Der Wal F., Lucas J., Silva C.S., Jourdain A., Muino J.M., Nanao M.H. The Intervening Domain Is Required for DNA-Binding and Functional Identity of Plant MADS Transcription Factors. Nat. Commun. 2021;12:4760. doi: 10.1038/s41467-021-24978-w. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

