Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation
- PMID: 39684695
- PMCID: PMC11641718
- DOI: 10.3390/ijms252312984
Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation
Abstract
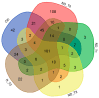
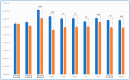
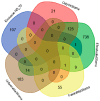
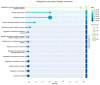
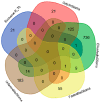
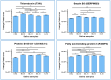
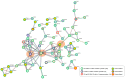
Glioblastoma multiforme (GBM) is an extremely aggressive brain tumor characterized by a high infiltration capability and recurrence rate. Early diagnosis is crucial to improve the prognosis and to personalize the therapeutic approach. This research explored, by LC-MS proteomic analysis after proteolytic digestion, the molecular profile of pre- and post-operative saliva pools from newly diagnosed (ND) GBM patients by comparing different times of collection and tumor recurrence (R). CYCS, PRDX2, RAB1C, PSMB1, KLK6, TMOD3, PAI2, PLBD1, CAST, and AHNAK, all involved in processes of tumor invasiveness and chemo- and radio-resistance, were found to depict the pre-surgery saliva of both ND and R GBM. PADI4 and CRYAB proteins, identified among the most abundant proteins exclusive of ND GBM pre-surgery saliva and classified as proteins elevated in glioma, could have a potential role as disease biomarkers. Selected panels of S100 proteins were found to potentially differentiate ND from R GBM patient saliva. TPD52 and IGKV3, exclusively identified in R GBM saliva, could be additionally distinctive of tumor relapse. Among the proteins identified in all pools, label-free relative quantitation showed statistically significant different levels of TXN, SERPINB5, FABP5, and S100A11 proteins between the pools. All of these proteins showed higher levels in both ND_ and R_T0 pre-surgery saliva with respect to CTRL and different modulation after surgery or chemo-radiotherapy combined treatment, suggesting a role as a potential panel of GBM predictive and prognostic biomarkers. These results highlight and confirm that saliva, a biofluid featured for an easily accessible and low invasiveness collection, is a promising source of GBM biomarkers, showing new potential opportunities for the development of targeted therapies and diagnostic tools.
Keywords: biomarkers; bottom-up proteomics; brain tumor; filter-aided sample preparation; glioblastoma multiforme; proteomics; saliva.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zhang W., Dang R., Liu H., Dai L., Liu H., Adegboro A.A., Zhang Y., Li W., Peng K., Hong J., et al. Machine learning-based investigation of regulated cell death for predicting prognosis and immunotherapy response in glioma patients. Sci. Rep. 2024;14:4173. doi: 10.1038/s41598-024-54643-3. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

