Characterization of an Activated Metabolic Transcriptional Program in Hepatoblastoma Tumor Cells Using scRNA-seq
- PMID: 39684755
- PMCID: PMC11641740
- DOI: 10.3390/ijms252313044
Characterization of an Activated Metabolic Transcriptional Program in Hepatoblastoma Tumor Cells Using scRNA-seq
Abstract
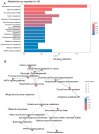
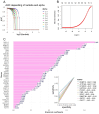
Hepatoblastoma is the most common primary liver malignancy in children, with metabolic reprogramming playing a critical role in its progression due to the liver's intrinsic metabolic functions. Enhanced glycolysis, glutaminolysis, and fatty acid synthesis have been implicated in hepatoblastoma cell proliferation and survival. In this study, we screened for altered overexpression of metabolic enzymes in hepatoblastoma tumors at tissue and single-cell levels, establishing and validating a hepatoblastoma tumor expression metabolic score using machine learning. Starting from the Mammalian Metabolic Enzyme Database, bulk RNA sequencing data from GSE104766 and GSE131329 datasets were analyzed using supervised methods to compare tumors versus adjacent liver tissue. Differential expression analysis identified 287 significantly regulated enzymes, 59 of which were overexpressed in tumors. Functional enrichment in the KEGG metabolic database highlighted a network enriched in amino acid metabolism, as well as carbohydrate, steroid, one-carbon, purine, and glycosaminoglycan metabolism pathways. A metabolic score based on these enzymes was validated in an independent cohort (GSE131329) and applied to single-cell transcriptomic data (GSE180665), predicting tumor cell status with an AUC of 0.98 (sensitivity 0.93, specificity 0.94). Elasticnet model tuning on individual marker expression revealed top tumor predictive markers, including FKBP10, ATP1A2, NT5DC2, UGT3A2, PYCR1, CKB, GPX7, DNMT3B, GSTP1, and OXCT1. These findings indicate that an activated metabolic transcriptional program, potentially influencing epigenetic functions, is observed in hepatoblastoma tumors and confirmed at the single-cell level.
Keywords: DNA methylation; cancer; epigenetics; hepatoblastoma; metabolism; one-carbon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Hager J., Sergi C.M. Hepatoblastoma. In: Departments of Pediatrics, Laboratory Medicine and Pathology, Stollery Children’s Hospital, University of Alberta, Edmonton, AB, Canada; Sergi C.M., editor. Liver Cancer. Exon Publications; Brisbane City, Australia: 2021. pp. 145–164.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

