Structures of Oligomeric States of Tau Protein, Amyloid-β, α-Synuclein and Prion Protein Implicated in Alzheimer's Disease, Parkinson's Disease and Prionopathies
- PMID: 39684761
- PMCID: PMC11641156
- DOI: 10.3390/ijms252313049
Structures of Oligomeric States of Tau Protein, Amyloid-β, α-Synuclein and Prion Protein Implicated in Alzheimer's Disease, Parkinson's Disease and Prionopathies
Abstract
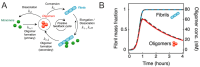
In this review, we focus on the biophysical and structural aspects of the oligomeric states of physiologically intrinsically disordered proteins and peptides tau, amyloid-β and α-synuclein and partly disordered prion protein and their isolations from animal models and human brains. These protein states may be the most toxic agents in the pathogenesis of Alzheimer's and Parkinson's disease. It was shown that oligomers are important players in the aggregation cascade of these proteins. The structural information about these structural states has been provided by methods such as solution and solid-state NMR, cryo-EM, crosslinking mass spectrometry, AFM, TEM, etc., as well as from hybrid structural biology approaches combining experiments with computational modelling and simulations. The reliable structural models of these protein states may provide valuable information for future drug design and therapies.
Keywords: amyloid-β; neurodegenerative diseases; oligomer; prion protein; tau protein; α-synuclein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Nguyen P.H., Ramamoorthy A., Sahoo B.R., Zheng J., Faller P., Straub J.E., Dominguez L., Shea J.-E., Dokholyan N.V., De Simone A., et al. Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer’s Disease, Parkinson’s Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis. Chem. Rev. 2021;121:2545–2647. doi: 10.1021/acs.chemrev.0c01122. - DOI - PMC - PubMed
-
- Nichols E., Steinmetz J.D., Vollset S.E., Fukutaki K., Chalek J., Abd-Allah F., Abdoli A., Abualhasan A., Abu-Gharbieh E., Akram T.T., et al. Estimation of the Global Prevalence of Dementia in 2019 and Forecasted Prevalence in 2050: An Analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7:e105–e125. doi: 10.1016/S2468-2667(21)00249-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

