AEmiGAP: AutoEncoder-Based miRNA-Gene Association Prediction Using Deep Learning Method
- PMID: 39684787
- PMCID: PMC11641653
- DOI: 10.3390/ijms252313075
AEmiGAP: AutoEncoder-Based miRNA-Gene Association Prediction Using Deep Learning Method
Abstract
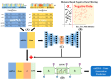
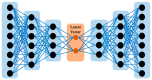
MicroRNAs (miRNAs) play a crucial role in gene regulation and are strongly linked to various diseases, including cancer. This study presents AEmiGAP, an advanced deep learning model that integrates autoencoders with long short-term memory (LSTM) networks to predict miRNA-gene associations. By enhancing feature extraction through autoencoders, AEmiGAP captures intricate, latent relationships between miRNAs and genes with unprecedented accuracy, outperforming all existing models in miRNA-gene association prediction. A thoroughly curated dataset of positive and negative miRNA-gene pairs was generated using distance-based filtering methods, significantly improving the model's AUC and overall predictive accuracy. Additionally, this study proposes two case studies to highlight AEmiGAP's application: first, a top 30 list of miRNA-gene pairs with the highest predicted association scores among previously unknown pairs, and second, a list of the top 10 miRNAs strongly associated with each of five key oncogenes. These findings establish AEmiGAP as a new benchmark in miRNA-gene association prediction, with considerable potential to advance both cancer research and precision medicine.
Keywords: LSTM; autoencoders; bioinformatics; cancer genomics; deep learning; feature extraction; miRNA–gene association; precision medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
SG-LSTM-FRAME: a computational frame using sequence and geometrical information via LSTM to predict miRNA-gene associations.Brief Bioinform. 2021 Mar 22;22(2):2032-2042. doi: 10.1093/bib/bbaa022. Brief Bioinform. 2021. PMID: 32181478
-
Identification of miRNA-disease associations via deep forest ensemble learning based on autoencoder.Brief Bioinform. 2022 May 13;23(3):bbac104. doi: 10.1093/bib/bbac104. Brief Bioinform. 2022. PMID: 35325038
-
MDFGNN-SMMA: prediction of potential small molecule-miRNA associations based on multi-source data fusion and graph neural networks.BMC Bioinformatics. 2025 Jan 13;26(1):13. doi: 10.1186/s12859-025-06040-4. BMC Bioinformatics. 2025. PMID: 39806287 Free PMC article.
-
LMI-DForest: A deep forest model towards the prediction of lncRNA-miRNA interactions.Comput Biol Chem. 2020 Dec;89:107406. doi: 10.1016/j.compbiolchem.2020.107406. Epub 2020 Oct 20. Comput Biol Chem. 2020. PMID: 33120126 Review.
-
Advances in applications of artificial intelligence algorithms for cancer-related miRNA research.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024 Apr 25;53(2):231-243. doi: 10.3724/zdxbyxb-2023-0511. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38650448 Free PMC article. Review. Chinese, English.
Cited by
-
Incorporating graph representation and mutual attention mechanism for MiRNA-MRNA interaction prediction.Front Genet. 2025 Jul 17;16:1637427. doi: 10.3389/fgene.2025.1637427. eCollection 2025. Front Genet. 2025. PMID: 40747104 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

