ADAM10 Expression by Ameloblasts Is Essential for Proper Enamel Formation
- PMID: 39684894
- PMCID: PMC11641948
- DOI: 10.3390/ijms252313184
ADAM10 Expression by Ameloblasts Is Essential for Proper Enamel Formation
Abstract
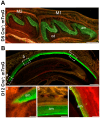
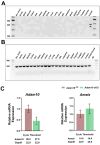
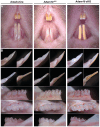
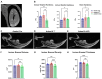
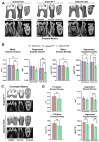
ADAM10 is a multi-functional proteinase that can cleave approximately 100 different substrates. Previously, it was demonstrated that ADAM10 is expressed by ameloblasts, which are required for enamel formation. The goal of this study was to determine if ADAM10 is necessary for enamel development. Deletion of Adam10 in mice is embryonically lethal and deletion of Adam10 from epithelia is perinatally lethal. We therefore deleted Adam10 from ameloblasts. Ameloblast-specific expression of the Tg(Amelx-iCre)872pap construct was confirmed. These mice were crossed with Adam10 floxed mice to generate Amelx-iCre; Adam10fl/fl mice (Adam10 cKO). The Adam10 cKO mice had discolored teeth with softer than normal enamel. Notably, the Adam10 cKO enamel density and volume were significantly reduced in both incisors and molars. Moreover, the incisor enamel rod pattern became progressively more disorganized, moving from the DEJ to the outer enamel surface, and this disorganized rod structure created gaps and S-shaped rods. ADAM10 cleaves proteins essential for cell signaling and for enamel formation such as RELT and COL17A1. ADAM10 also cleaves cell-cell contacts such as E- and N-cadherins that may support ameloblast movement necessary for normal rod patterns. This study shows, for the first time, that ADAM10 expressed by ameloblasts is essential for proper enamel formation.
Keywords: Cre Lox recombination; ameloblast migration; cervical enamel; density; hardness; mouse; occlusal enamel; volume.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

