A Role for Periostin Pathological Variants and Their Interaction with HSP70-1a in Promoting Pancreatic Cancer Progression and Chemoresistance
- PMID: 39684914
- PMCID: PMC11641934
- DOI: 10.3390/ijms252313205
A Role for Periostin Pathological Variants and Their Interaction with HSP70-1a in Promoting Pancreatic Cancer Progression and Chemoresistance
Abstract
Pancreatic ductal adenocarcinoma (PDAC) characterized by an abundant cancer stroma is an aggressive malignancy with a poor prognosis. Periostin (Pn) is a key extracellular matrix (ECM) protein in various tumor progression. Previously, we described the role of Pn alternative splicing variants (ASVs) with specific functional features in breast cancer. Pn is known to associate with a chemoresistance of PDAC, but the functions of the Pn-ASVs remain largely unknown. In this study, we focused on physiological and pathological Pn-ASVs, and examined the characteristics of Pn-expressing cells and the difference in function of each ASV. We found that cancer-associated fibroblasts (CAFs) are a main source of Pn synthesis, which selectively secrete pathological Pn-ASVs with exon 21 both in mouse and human samples. RNA sequencing identified a gene signature of Pn-positive CAFs associated with ECM-related genes and chemokines, factors that shape the chemoresistance tumor microenvironment (TME). Additionally, only pathological Pn-ASVs interacted with heat shock protein 70-1a (HSP70-1a), leading to significant rescue of gemcitabine-induced PDAC apoptosis. In silico analysis revealed that the presence or absence of exon 21 changes the tertiary structure of Pn and the binding sites for HSP70-1a. Altogether, Pn-ASVs with exon 21 secreted from CAFs play a key role in supporting tumor growth by interacting with cancer cell-derived HSP70-1a, indicating that Pn-ASVs with exon 21 might be a potential therapeutic and diagnostic target in PDAC patients with rich stroma.
Keywords: alternative splicing variants; extracellular matrix protein; pancreatic cancers; periostin.
Conflict of interest statement
R. Morishita received honoraria, consulting fees and funds from Novartis, Takeda, Shionogi, Astellas, Boehringer Ingelheim, Daiichi-Sankyo and Pfizer. F. Sanada, Y. Taniyama and K. Shibata are members of Periotherapia Co., Ltd.
Figures
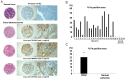

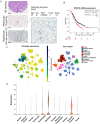


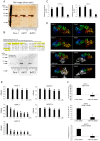
References
-
- Neuzillet C., Nicolle R., Raffenne J., Tijeras-Raballand A., Brunel A., Astorgues-Xerri L., Vacher S., Arbateraz F., Fanjul M., Hilmi M., et al. Periostin- and podoplanin-positive cancer-associated fibroblast subtypes cooperate to shape the inflamed tumor microenvironment in aggressive pancreatic adenocarcinoma. J. Pathol. 2022;258:408–425. doi: 10.1002/path.6011. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

