Colposcopy referrals and CIN3 detection after triage by host cell DNA methylation and/or HPV genotyping in HPV positive women with low-grade cytology from a population-based Dutch primary HPV screening trial
- PMID: 39686532
- PMCID: PMC11701402
- DOI: 10.1002/ijc.35289
Colposcopy referrals and CIN3 detection after triage by host cell DNA methylation and/or HPV genotyping in HPV positive women with low-grade cytology from a population-based Dutch primary HPV screening trial
Abstract
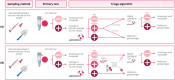
High-risk HPV (hrHPV)-based screening has led to many unnecessary colposcopy referrals, mainly because of direct referral after low-grade cytology (ASC-US/LSIL). DNA methylation and genotyping tests on ASC-US/LSIL samples have the potential to significantly improve the efficiency of screening. In this study, 12 triage strategies were constructed from FAM19A4/miR124-2 or ASCL1/LHX8 methylation, HPV16/18 or HPV16/18/31/33/45 genotyping and 1-year repeat cytology. The performance was evaluated on 215 hrHPV-positive ASC-US/LSIL samples from the IMPROVE trial (NTR5078). Performance was measured by colposcopy referral rate, positive predictive value (PPV) for detecting precancer (CIN3), and negative predictive value (NPV). To evaluate efficiency, strategies were ordered by the cumulative colposcopy referral rate after 1-year cytology and compared by the marginal PPV to detect one additional CIN3 (mPPV). The most conservative strategy (referral when HPV16/18 and FAM19A4/miR124 methylation results are positive) had a direct referral rate of 5.2%, a cumulative referral rate after 1-year cytology of 54.1%, and mPPV of 19.3%. Replacing HPV16/18 by HPV16/18/31/33/45 increased the cumulative 1-year referral rate to 54.6%, and yielded an mPPV of 10.0%. Similar results were obtained for strategies with ASCL1/LHX8 methylation. Of all strategies, referral after an HPV16/18/31/33/45 positive, ASCL1/LHX8 methylation-positive, and/or 1-year cytology-positive result yielded the highest direct and cumulative 1-year colposcopy referral rates of 64.4% and 79.1%, respectively. The NPVs after 1-year cytology varied between 98.1% and 99.4%, warranting a return to routine screening. Altogether, DNA methylation-based triage strategies are recommended as they are discriminative for CIN3 and control the number of immediate colposcopy referrals.
Keywords: ASC‐US/LSIL; DNA methylation; cervical cancer; colposcopy referrals; diagnostic markers.
© 2024 The Author(s). International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Conflict of interest statement
DAMH, RDMS, and CJLMM are minority shareholders of Self‐screen B.V., a spin‐off company of VUmc (currently known as AmsterdamUMC location Vrije Universiteit Amsterdam); Self‐screen B.V. develops, manufactures, and licenses high‐risk HPV and methylation marker assays for cervical cancer screening and holds patents on these tests; CJLMM is part‐time CEO of Self‐screen B.V., and served occasionally on the scientific advisory boards of Qiagen; the other authors declare no conflicts of interests.
Figures

References
-
- Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30(Suppl 5):F88‐F99. - PubMed
-
- Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV‐based screening for prevention of invasive cervical cancer: follow‐up of four European randomised controlled trials. Lancet. 2014;383(9916):524‐532. - PubMed
-
- Machalek DA, Roberts JM, Garland SM, et al. Routine cervical screening by primary HPV testing: early findings in the renewed National Cervical Screening Program. Med J Aust. 2019;211(3):113‐119. - PubMed
-
- Maver PJ, Poljak M. Primary HPV‐based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;26(5):579‐583. - PubMed
-
- Loopik DL, Koenjer LM, Siebers AG, Melchers WJG, Bekkers RLM. Benefit and burden in the Dutch cytology‐based vs high‐risk human papillomavirus‐based cervical cancer screening program. Am J Obstet Gynecol. 2021;224(2):200.e1‐200.e9. - PubMed

