Less, but More: New Insights From Appendicularians on Chordate Fgf Evolution and the Divergence of Tunicate Lifestyles
- PMID: 39686543
- PMCID: PMC11733497
- DOI: 10.1093/molbev/msae260
Less, but More: New Insights From Appendicularians on Chordate Fgf Evolution and the Divergence of Tunicate Lifestyles
Abstract
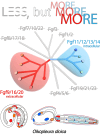
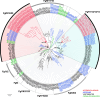
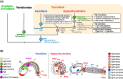
The impact of gene loss on the diversification of taxa and the emergence of evolutionary innovations remains poorly understood. Here, our investigation on the evolution of the Fibroblast Growth Factors (FGFs) in appendicularian tunicates as a case study reveals a scenario of "less, but more" characterized by massive losses of all Fgf gene subfamilies, except for the Fgf9/16/20 and Fgf11/12/13/14, which in turn underwent two bursts of duplications. Through phylogenetic analysis, synteny conservation, and gene and protein structure, we reconstruct the history of appendicularian Fgf genes, highlighting their paracrine and intracellular functions. An exhaustive analysis of developmental Fgf expression in Oikopleura dioica allows us to identify four associated evolutionary patterns characterizing the "less, but more" conceptual framework: conservation of ancestral functions; function shuffling between paralogs linked to gene losses; innovation of new functions after the duplication bursts; and function extinctions linked to gene losses. Our findings allow us to formulate novel hypotheses about the impact of Fgf losses and duplications on the transition from an ancestral ascidian-like biphasic lifestyle to the fully free-living appendicularians. These hypotheses include massive co-options of Fgfs for the development of the oikoblast and the tail fin; recruitment of Fgf11/12/13/14s into the evolution of a new mouth, and their role modulating neuronal excitability; the evolutionary innovation of an anterior tail FGF signaling source upon the loss of retinoic acid signaling; and the potential link between the loss of Fgf7/10/22 and Fgf8/17/18 and the loss of drastic metamorphosis and tail absorption in appendicularians, in contrast to ascidians.
Keywords: Oikopleura dioica; FGF signaling; alternative splicing; appendicularian; embryo development; gene duplication; gene loss; tunicate evolution; “less, but more”.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Massive Gene Loss and Function Shuffling in Appendicularians Stretch the Boundaries of Chordate Wnt Family Evolution.Front Cell Dev Biol. 2021 Jun 9;9:700827. doi: 10.3389/fcell.2021.700827. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34179025 Free PMC article.
-
Cardiopharyngeal deconstruction and ancestral tunicate sessility.Nature. 2021 Nov;599(7885):431-435. doi: 10.1038/s41586-021-04041-w. Epub 2021 Nov 17. Nature. 2021. PMID: 34789899
-
Functional evolutionary history of the mouse Fgf gene family.Dev Dyn. 2008 Jan;237(1):18-27. doi: 10.1002/dvdy.21388. Dev Dyn. 2008. PMID: 18058912 Review.
-
Evolutionary diversification of secondary mechanoreceptor cells in tunicata.BMC Evol Biol. 2013 Jun 4;13:112. doi: 10.1186/1471-2148-13-112. BMC Evol Biol. 2013. PMID: 23734698 Free PMC article.
-
The simple tail of chordates: phylogenetic significance of appendicularians.Genesis. 2001 Jan;29(1):36-45. doi: 10.1002/1526-968x(200101)29:1<36::aid-gene1003>3.0.co;2-j. Genesis. 2001. PMID: 11135460 Review.
Cited by
-
Unique trajectory of gene family evolution from genomic analysis of nearly all known species in an ancient yeast lineage.Mol Syst Biol. 2025 Aug;21(8):1066-1089. doi: 10.1038/s44320-025-00118-0. Epub 2025 May 27. Mol Syst Biol. 2025. PMID: 40425814 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

