Lonafarnib Protects Against Muscle Atrophy Induced by Dexamethasone
- PMID: 39686867
- PMCID: PMC11696026
- DOI: 10.1002/jcsm.13665
Lonafarnib Protects Against Muscle Atrophy Induced by Dexamethasone
Abstract
Background: Muscle atrophy, including glucocorticoid-induced muscle wasting from treatments such as dexamethasone (DEX), results in significant reductions in muscle mass, strength and function. This study investigates the potential of lonafarnib, a farnesyltransferase inhibitor, to counteract DEX-induced muscle atrophy by targeting key signalling pathways.
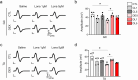
Methods: We utilized in vitro models with C2C12 myotubes treated with DEX and in vivo models with Caenorhabditis elegans and DEX-treated Sprague-Dawley rats. Myotube morphology was assessed by measuring area, fusion index and diameter. Muscle function was evaluated by grip strength and compound muscle action potential (CMAP) in the gastrocnemius (GC) and tibialis anterior (TA) muscles. Molecular mechanisms were explored through RNA sequencing and Western blotting to assess changes in mitochondrial function and muscle signalling pathways.
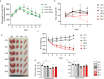
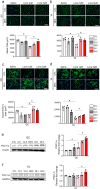
Results: Lonafarnib (2 μM) significantly improved myotube area (1.49 ± 0.14 × 105 μm2 vs. 1.03 ± 0.49 × 105 μm2 in DEX, p < 0.05), fusion index (18.73 ± 1.23% vs. 13.3 ± 1.56% in DEX, p < 0.05) and myotube diameter (31.89 ± 0.89 μm vs. 21.56 ± 1.01 μm in DEX, p < 0.05) in C2C12 myotubes. In C. elegans, lonafarnib (100 μM) increased the pharyngeal pumping rate from 212 ± 7.21 contractions/min in controls to 308 ± 17.09 contractions/min at day 4 (p < 0.05), indicating enhanced neuromuscular function. In DEX-induced atrophic rats, lonafarnib improved maximal grip strength (DEX: 13.91 ± 0.78 N vs. 1 μM lonafarnib: 16.18 ± 0.84 N and 5 μM lonafarnib: 16.71 ± 0.83 N, p < 0.05), increased muscle weight in GC, and enhanced CMAP amplitudes in both GC and TA muscles. Western blot analysis showed that lonafarnib treatment upregulated UCP3 and ANGPTL4 and increased phosphorylation of mTOR and S6 ribosomal protein (p < 0.05), indicating enhanced mitochondrial function and protein synthesis. Knockdown models further demonstrated that lonafarnib could partially rescue muscle atrophy phenotypes, indicating its action through multiple molecular pathways.
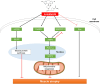
Conclusions: Lonafarnib mitigates dexamethasone-induced muscle atrophy by enhancing mitochondrial function and activating anabolic pathways. These findings support further investigation of lonafarnib as a therapeutic agent for muscle atrophy in clinical settings.
Keywords: ANGPLT4; UCP3; dexamethasone; drug repositioning; lonafarnib; muscle atrophy.
© 2025 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Cruz‐Jentoft A. J. and Sayer A. A., “Sarcopenia,” Lancet 393 (2019): 2636–2646. - PubMed
-
- Suzuki M., Jeng L. J. B., Chefo S., et al., “FDA Approval Summary for Lonafarnib (Zokinvy) for the Treatment of Hutchinson‐Gilford Progeria Syndrome and Processing‐Deficient Progeroid Laminopathies,” Genetics in Medicine 25 (2023): 100335. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

