Blastocyst complementation-based rat-derived heart generation reveals cardiac anomaly barriers to interspecies chimera development
- PMID: 39687030
- PMCID: PMC11647242
- DOI: 10.1016/j.isci.2024.111414
Blastocyst complementation-based rat-derived heart generation reveals cardiac anomaly barriers to interspecies chimera development
Abstract
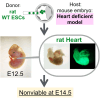
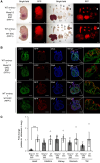
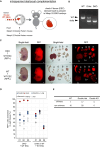
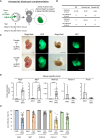
The use of pluripotent stem cells (PSCs) to generate functional organs via blastocyst complementation is a cutting-edge strategy in regenerative medicine. However, existing models that use this method for heart generation do not meet expectations owing to the complexity of heart development. Here, we investigated a Mesp1/2 deficient mouse model, which is characterized by abnormalities in the cardiac mesodermal cells. The injection of either mouse or rat PSCs into Mesp1/2 deficient mouse blastocysts led to successful heart generation. In chimeras, the resulting hearts were predominantly composed of rat cells; however, their functionality was limited to the embryonic developmental stage on day 12.5. These results present the functional limitation of the xenogeneic heart, which poses a significant challenge to the development in mouse-rat chimeras.
Keywords: Health sciences; biological sciences; cardiovascular medicine.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing or financial interests.
Figures



References
LinkOut - more resources
Full Text Sources

