RNPS1 in PSAP complex controls periodic pre-mRNA splicing over the cell cycle
- PMID: 39687031
- PMCID: PMC11648250
- DOI: 10.1016/j.isci.2024.111400
RNPS1 in PSAP complex controls periodic pre-mRNA splicing over the cell cycle
Abstract
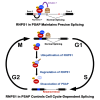
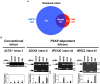
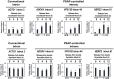
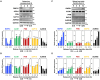
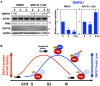
Cell cycle progression requires periodic gene expression through splicing control. However, the splicing factor that directly controls this cell cycle-dependent splicing remains unknown. Cell cycle-dependent expression of the AURKB (aurora kinase B) gene is essential for chromosome segregation and cytokinesis. We previously reported that RNPS1 is essential to maintain precise splicing in AURKB intron 5. Here we show that RNPS1 plays this role in PSAP complex with PNN and SAP18, but not ASAP complex with ACIN1 and SAP18. Whole-transcriptome sequencing of RNPS1- and PNN-deficient cells indicated that RNPS1, either alone or as PSAP complex, is an essential splicing factor for a subset of introns. Remarkably, protein expression of RNPS1, but not PNN, is coordinated with cyclical splicing in PSAP-controlled introns including AURKB intron 5. The ubiquitin-proteasome pathway is involved in the periodic decrease of RNPS1 protein level. RNPS1 is a key factor that controls periodic splicing during the cell cycle.
Keywords: Cell biology; Cellular physiology; Molecular biology; Properties of biomolecules.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

