Risk of intraoperative hemorrhage during cesarean scar ectopic pregnancy surgery: development and validation of an interpretable machine learning prediction model
- PMID: 39687425
- PMCID: PMC11646795
- DOI: 10.1016/j.eclinm.2024.102969
Risk of intraoperative hemorrhage during cesarean scar ectopic pregnancy surgery: development and validation of an interpretable machine learning prediction model
Abstract
Background: Current models for predicting intraoperative hemorrhage in cesarean scar ectopic pregnancy (CSEP) are constrained by known risk factors and conventional statistical methods. Our objective is to develop an interpretable prediction model using machine learning (ML) techniques to assess the risk of intraoperative hemorrhage during CSEP in women, followed by external validation and clinical application.
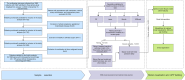
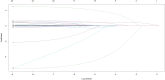
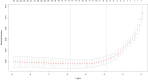
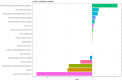
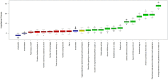
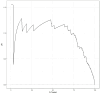
Methods: This multicenter retrospective study utilized electronic medical record (EMR) data from four tertiary medical institutions. The model was developed using data from 1680 patients with CSEP diagnosed and treated at Qilu Hospital of Shandong University, Chongqing Health Center for Women and Children, and Dezhou Maternal and Child Health Care Hospital between January 1, 2008, and December 31, 2023. External validation data were obtained from Liao Cheng Dong Chang Fu District Maternal and Child Health Care Hospital between January 1, 2021, and December 31, 2023. Random forest (RF), Lasso, Boruta, and Extreme Gradient Boosting (XGBoost) were employed to identify the most influential variables in the model development data set; the best variables were selected based on reaching the λmin value. Model development involved eight machine learning methods with ten-fold cross-validation. Accuracy and decision curve analysis (DCA) were used to assess model performance for selection of the optimal model. Internal validation of the model utilized area under the receiver operating characteristic curve (AUC), sensitivity, specificity, Matthews correlation coefficient, and F1 score. These same indicators were also applied to evaluate external validation performance of the model. Finally, visualization techniques were used to present the optimal model which was then deployed for clinical application via network applications.
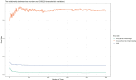
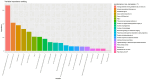
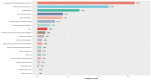
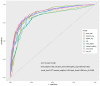
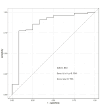
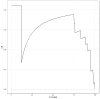
Findings: Setting λmin at the value of 0.003, the optimal variable combination containing 9 variables was selected for model development. The optimal prediction model (Bayes) had an accuracy of 0.879 (95% CI: 0.857-0.901) an AUC of 0.882 (95% CI: 0.860-0.904), a DCA curve maximum threshold probability of 0.41, and a maximum return of 7.86%. The internal validation accuracy was 0.869 (95% CI: 0.847-0.891), an AUC of 0.822 (95% CI: 0.801-0.843), a sensitivity of 0.938, a specificity of 0.422, a Matthews correlation coefficient of 0.392, and an F1 score of 0.925. In the external validation, the accuracy was 0.936 (95% CI: 0.913-0.959), an AUC of 0.853 (95% CI: 0.832-0.874), a sensitivity of 0.954, a specificity of 0.5, a Matthews correlation coefficient of 0.365, and an F1 score of 0.966. This indicates that the prediction model performed well in both internal and external validation.
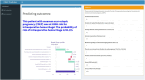
Interpretation: The developed prediction model, deployed in the network application, is capable of forecasting the risk of intraoperative hemorrhage during CSEP. This tool can facilitate targeted preoperative assessment and clinical decision-making for clinicians. Prospective data should be utilized in future studies to further validate the extended applicability of the model.
Funding: Natural Science Foundation of Shandong Province; Qilu Hospital of Shandong University.
Keywords: Cesarean scar ectopic pregnancy; Interpretable machine learning; Intraoperative hemorrhage; Prediction model; Visualization.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest related to this work.
Figures











References
-
- Liu L., Ross W.T., Chu A.L., Deimling T.A. An updated guide to the diagnosis and management of cesarean scar pregnancies. Curr Opin Obstet Gynecol. 2020;32(4):255–262. - PubMed
-
- Tan K.L., Jiang L., Chen Y.M., et al. Local intra-gestational sac methotrexate injection followed by dilation and curettage in treating cesarean scar pregnancy. Arch Gynecol Obstet. 2020;302(2):439–445. - PubMed
-
- De Braud L.V., Knez J., Mavrelos D., Thanatsis N., Jauniaux E., Jurkovic D. Risk prediction of major haemorrhage with surgical treatment of live cesarean scar pregnancies. Eur J Obstet Gynecol Reprod Biol. 2021;264:224–231. - PubMed
-
- Yang F., Yang X., Jing H., et al. MRI-based scoring model to predict massive hemorrhage during dilatation and curettage in patients with cesarean scar pregnancy. Abdom Radiol (NY) 2023;48(10):3195–3206. - PubMed
LinkOut - more resources
Full Text Sources

