Utilizing an In-silico Approach to Pinpoint Potential Biomarkers for Enhanced Early Detection of Colorectal Cancer
- PMID: 39687502
- PMCID: PMC11648020
- DOI: 10.1177/11769351241307163
Utilizing an In-silico Approach to Pinpoint Potential Biomarkers for Enhanced Early Detection of Colorectal Cancer
Abstract
Objectives: Colorectal cancer (CRC) is a prevalent disease characterized by significant dysregulation of gene expression. Non-invasive tests that utilize microRNAs (miRNAs) have shown promise for early CRC detection. This study aims to determine the association between miRNAs and key genes in CRC.
Methods: Two datasets (GSE106817 and GSE23878) were extracted from the NCBI Gene Expression Omnibus database. Penalized logistic regression (PLR) and artificial neural networks (ANN) were used to identify relevant miRNAs and evaluate the classification accuracy of the selected miRNAs. The findings were validated through bipartite miRNA-mRNA interactions.
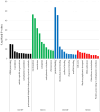
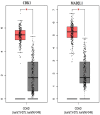
Results: Our analysis identified 3 miRNAs: miR-1228, miR-6765-5p, and miR-6787-5p, achieving a total accuracy of over 90%. Based on the results of the mRNA-miRNA interaction network, CDK1 and MAD2L1 were identified as target genes of miR-6787-5p.
Conclusions: Our results suggest that the identified miRNAs and target genes could serve as non-invasive biomarkers for diagnosing colorectal cancer, pending laboratory confirmation.
Keywords: Colorectal neoplasms; artificial neural networks; least absolute shrinkage and selection operator; microRNA; smoothly clipped absolute deviation; the minimax concave penalty.
© The Author(s) 2024.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
-
- Morgan E, Arnold M, Gini A, et al. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-344. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

