Spatial transcriptomics and in situ immune cell profiling of the host ectocervical landscape of HIV infected Kenyan sex working women
- PMID: 39687623
- PMCID: PMC11646855
- DOI: 10.3389/fimmu.2024.1483346
Spatial transcriptomics and in situ immune cell profiling of the host ectocervical landscape of HIV infected Kenyan sex working women
Abstract
Introduction: Chronic immune activation is a hallmark of human immunodeficiency virus (HIV) infection that significantly impacts disease pathogenesis. However, in-depth studies characterizing the immunological landscape of the ectocervix during chronic HIV infection remain scarce despite the importance of this tissue site for HIV transmission.
Methods: Ectocervical tissue samples were obtained from antiretroviral-naïve HIV-seropositive and -seronegative Kenyan female sex workers. These samples were assessed by spatial transcriptomics and Gene Set Enrichment Analysis. We further performed multi-epitope ligand cartography (MELC) using an in situ staining panel that included 17 markers of primarily T cell-mediated immune responses.
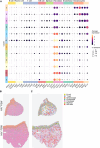
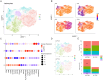
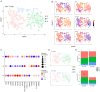
Results: Spatial transcriptomics revealed tissue-wide immune activation encompassing immune responses associated with chronic HIV infection. First, both the epithelial and submucosal compartments showed diverse but significant upregulation of humoral immune responses, as indicated by the expression of several antibody-related genes. Second, an antiviral state-associated cellular immunity was also observed in the HIV-seropositive group, characterized by upregulation of genes involved in interferon signaling across the mucosal tissue and a more spatially restricted mucosal expression of genes related to T cell activity and effector functions relative to the HIV-seronegative group. Additionally, HIV associated structural alterations were evident within both compartments. Downregulated genes across the epithelium were mainly linked to epithelial integrity, with the outer layer involved in terminal differentiation and the inner layer associated with epithelial structure. MELC analysis further revealed a significantly increased ectocervical leukocyte population in HIV-seropositive participants, primarily driven by an increase in CD8+ T cells while the CD4+ T cell population remained stable. Consistent with our spatial transcriptomics data, T cells from HIV-seropositive participants showed an increased effector phenotype, defined by elevated expression of various granzymes.
Conclusion: By combining spatial transcriptomics and MELC, we identified significant HIV-associated cervical immune activity driven by induction of both T and B cell activity, together with a general antiviral state characterized by sustained interferon induction. These findings underscore that chronic HIV infection is associated with an altered ectocervical mucosal immune landscape years after primary infection. This sheds light on HIV pathogenesis at distant local sites and complements current knowledge on HIV-associated systemic immune activation.
Keywords: B cells; HIV; T cells; interferon; mucosal immunology; multi-epitope ligand cartography; spatial transcriptomics.
Copyright © 2024 Franzén Boger, Kaldhusdal, Pascual-Reguant, Kroh, Uecker, Burgener, Lajoie, Omollo, Kimani, Fowke, Hauser, Tjernlund and Broliden.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- So-Armah KA, Tate JP, Chang CCH, Butt AA, Gerschenson M, Gibert CL, et al. Do biomarkers of inflammation, monocyte activation, and altered coagulation explain excess mortality between HIV infected and uninfected people? J Acquir Immune Defic Syndr. (2016) 72:206. doi: 10.1097/QAI.0000000000000954 - DOI - PMC - PubMed
-
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. (2006) 12:1365–71. https://www.nature.com/articles/nm1511. - PubMed
-
- Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. (1999) 179:859–70. doi: 10.1086/314660 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

