mRNA vaccines encoding membrane-anchored RBDs of SARS-CoV-2 mutants induce strong humoral responses and can overcome immune imprinting
- PMID: 39687732
- PMCID: PMC11646785
- DOI: 10.1016/j.omtm.2024.101380
mRNA vaccines encoding membrane-anchored RBDs of SARS-CoV-2 mutants induce strong humoral responses and can overcome immune imprinting
Abstract
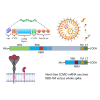
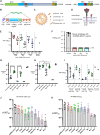
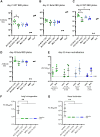
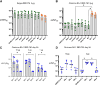
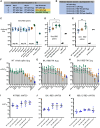
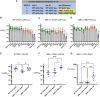
We investigated mRNA vaccines encoding a membrane-anchored receptor-binding domain (RBD), each a fusion of a variant RBD, the transmembrane (TM) and cytoplasmic tail fragments of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. In naive mice, RBD-TM mRNA vaccines against SARS-CoV-2 variants induced strong humoral responses against the target RBD. Multiplex surrogate viral neutralization (sVNT) assays revealed broad neutralizing activity against a range of variant RBDs. In the setting of a heterologous boost, against the background of exposure to ancestral whole-spike vaccines, sVNT studies suggested that BA.1 and BA.5 RBD-TM vaccines had the potential to overcome the detrimental effects of immune imprinting. A subsequent heterologous boost study using XBB.1.5 booster vaccines was evaluated using both sVNT and authentic virus neutralization. Geometric mean XBB.1.5 neutralization values after third-dose RBD-TM or whole-spike XBB.1.5 booster vaccines were compared with those after a third dose of ancestral spike booster vaccine. Fold-improvement over ancestral vaccine was just 1.3 for the whole-spike XBB.1.5 vaccine, similar to data published using human serum samples. In contrast, the fold-improvement achieved by the RBD-TM XBB.1.5 vaccine was 16.3, indicating that the RBD-TM vaccine induced the production of antibodies that neutralize the XBB.1.5 variant despite previous exposure to ancestral spike protein.
Keywords: COVID vaccine; anchored RBD vaccine; immune imprinting; lipid nanoparticle; mRNA vaccine; virus neutralization.
Crown Copyright © 2024 Published by Elsevier Inc. on behalf of The American Society of Gene and Cell Therapy.
Conflict of interest statement
Two provisional patents (PCT/AU2022/050912 and PCT/AU2022/050913) covering the RBD-TM mRNA vaccine design and the LNP formulation used in this study, and underlying technology, have been submitted through Monash University, with C.W.P., H.A.W., and S.A.F. as co-inventors of 050912 and C.W.P., H.A.W., and J.K.H. as co-inventors of 050913. C.W.T. and L.-F.W. are co-inventors of a patent on the surrogate VNT test (sVNT) platform. T.N. receives research contracts to conduct clinical trials, with funding to institutions from Moderna, SanofiPasteur, GSK, Iliad Biotechnologies, Dynavax, Seqirus, Janssen, and MSD. T.N. receives consulting fees from GSK, Seqirus, MSD, SanofiPasteur, AstraZeneca, Moderna, BioNet, and Pfizer. T.N. serves on DSMBs for Seqirus, Clover, Moderna, Emergent, Serum Institute of India, SK Bioscience Korea, Emergent Biosolutions, and Novavax. S.R. is an employee of CSL Seqirus that is a maker of influenza vaccines. C.Y.W. is a shareholder of Ena Respiratory. D.I.G. has received research funding from CSL for an unrelated project.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

