Assessment of genetic structure and trait associations of Watkins wheat landraces under Egyptian field conditions
- PMID: 39687740
- PMCID: PMC11646717
- DOI: 10.3389/fgene.2024.1384220
Assessment of genetic structure and trait associations of Watkins wheat landraces under Egyptian field conditions
Abstract
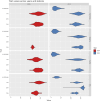
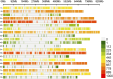
Background: Wheat landraces represent a reservoir of genetic diversity that can support wheat improvement through breeding. A core panel of 300 Watkins wheat landraces, as well as 16 non-Watkins landraces and elite wheat cultivars, was grown during the 2020-2021 and 2021-2022 seasons at four Agricultural Research Stations in Egypt, Gemmiza, Nubaria, Sakha, and Sids, to evaluate the core panel for agromorphological and yield-related traits. The genetic population structure within these genotypes were assessed using 35,143 single nucleotide polymorphisms (SNPs).
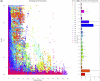
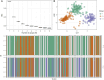
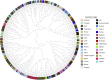
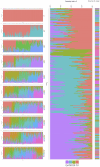
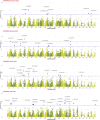
Results: Cluster analyses using Discriminant Analysis of Principal Components (DAPC) and k-means revealed three clusters with moderate genetic differentiation and population structure, possibly due to wheat breeding systems and geographical isolation. The best ancestry was k = 4, but k = 2 and k = 3 were also significant. A genome-wide association study (GWAS) identified clustered marker trait associations (MTAs) linked to thousand kernel weight on chromosome 5A, plant height on chromosomes 3B and 1D, days to heading on chromosomes 2A, 4B, 5B and 1D, and plant maturity on chromosomes 3A, 2B, and 6B. In the future, these MTAs can be used to accelerate the incorporation of beneficial alleles into locally adapted germplasm through marker-assisted selection. Gene enrichment analysis identified key genes within these loci, including Reduced height-1 (Rht-A1) and stress-related genes.
Conclusion: These findings underscore significant genetic connections and the involvement of crucial biological pathways.
Keywords: Watkins; agromorphological traits; genome-wide association study (GWAS); marker trait associations; population structure; wheat.
Copyright © 2024 Elkot, Nassar, Elmassry, Forner-Martínez, Awal, Wingen, Griffiths, Alsamman and Kehel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Aalto S., Kiiras K., Mayer H., Miettinen H. (2022). Armed conflict in Ukraine. Perspect. Geogr. inequalities Sustain. Dev. Goals—five case Stud. 42. 10.1016/j.geosus.2022.09.003 - DOI
-
- Abay K. (2023). Wheat genetics, research and development in Egypt. International Food Policy Research Institute.
-
- Abdalla A., Becker M., Stellmacher T. (2023). The contribution of agronomic management to sustainably intensify Egypt’s wheat production. Agriculture 13, 978. 10.3390/agriculture13050978 - DOI
-
- Abdelmageed K., Chang X.-h., Wang D.-m., Wang Y.-j., Yang Y.-s., Zhao G.-c., et al. (2019). Evolution of varieties and development of production technology in Egypt wheat: a review. J. Integr. Agric. 18, 483–495. 10.1016/s2095-3119(18)62053-2 - DOI
-
- Aleksandrov V., Kartseva T., Alqudah A. M., Kocheva K., Tasheva K., Börner A., et al. (2021). Genetic diversity, linkage disequilibrium and population structure of Bulgarian bread wheat assessed by genome-wide distributed snp markers: from old germplasm to semi-dwarf cultivars. Plants 10, 1116. 10.3390/plants10061116 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

