METTL4-Mediated Mitochondrial DNA N6-Methyldeoxyadenosine Promoting Macrophage Inflammation and Atherosclerosis
- PMID: 39687989
- PMCID: PMC11952693
- DOI: 10.1161/CIRCULATIONAHA.124.069574
METTL4-Mediated Mitochondrial DNA N6-Methyldeoxyadenosine Promoting Macrophage Inflammation and Atherosclerosis
Abstract
Background: Mitochondrial dysfunction is a key factor in the development of atherogenesis. METTL4 (methyltransferase-like protein 4) mediates N6- methyldeoxyadenosine (6mA) of mammalian mitochondrial DNA (mtDNA). However, the role of METTL4-mediated mitoepigenetic regulation in atherosclerosis is still unknown. This study aims to investigate the potential involvement of METTL4 in atherosclerosis, explore the underlying mechanism, and develop targeted strategies for treating atherosclerosis.
Methods: Expression levels of mtDNA 6mA and METTL4 were determined in atherosclerotic lesions. We explored the mechanism of METTL4 involvement in atherosclerosis using Mettl4Mac-KO-Apoe-/- and Mettl4MUT-Apoe-/- mice and cell models, as well as bone marrow transplantation. Natural compound libraries were screened to identify potent METTL4 antagonists. In addition, bioinspired proteolysis targeting chimera technology targeting macrophages within plaques was used to increase the efficacy of the METTL4 antagonist.
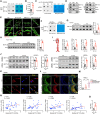
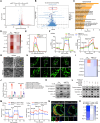
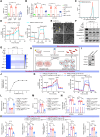
Results: The expression levels of mtDNA 6mA and METTL4 were significantly increased in plaque macrophages. Mettl4Mac-KO-Apoe-/- mice displayed suppressed mtDNA 6mA levels and atherosclerotic progression, which were reversed by METTL4 restoration through bone marrow transplantation (n=6). Mechanistically, elevated METTL4 expression reduces mitochondrial ATP6 (MT-ATP6) expression by suppressing its transcription, thereby impairing the activity of mitochondrial respiration chain complex V. This disruption leads to the accumulation of excess protons in the mitochondrial intermembrane space, causing mitochondrial dysfunction. Consequently, mtDNA is released into the cytoplasm, ultimately triggering inflammasome activation. All results were reversed by the mutation in the METTL4 methyltransferase active site. Mettl4MUT-Apoe-/- mice showed suppressed mtDNA 6mA levels and atherosclerotic progression and repaired mitochondrial function of macrophage, which were reversed by METTL4 restoration through bone marrow transplantation (n=6). Pemetrexed was identified as the first METTL4 antagonist to effectively alleviate atherosclerotic progression. Furthermore, we generated a proteolysis targeting chimera drug based on pemetrexed that specifically targeted METTL4 in macrophages within plaques, showing a promising therapeutic effect on atherosclerosis.
Conclusions: This study revealed a novel mechanism by which mtDNA 6mA orchestrated mitochondrial function-related gene expression in macrophages, thereby promoting atherosclerosis. Through various experimental techniques, such as gene manipulation, pharmacological inhibition, and proteolysis targeting chimera, this study demonstrated that mtDNA 6mA and its specific enzyme METTL4 hold potential as therapeutic targets for atherosclerosis.
Keywords: METTL4; atherosclerosis; inflammation; macrophages; mitochondria.
Conflict of interest statement
None.
Figures





References
-
- Zhu D, Li X, Tian Y. Mitochondrial-to-nuclear communication in aging: an epigenetic perspective. Trends Biochem Sci. 2022;47:645–659. doi: 10.1016/j.tibs.2022.03.008 - PubMed
-
- Madamanchi NR, Runge MS. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100:460–473. doi: 10.1161/01.RES.0000258450.44413.96 - PubMed
-
- Dumont A, Lee M, Barouillet T, Murphy A, Yvan-Charvet L. Mitochondria orchestrate macrophage effector functions in atherosclerosis. Mol Aspects Med. 2021;77:100922. doi: 10.1016/j.mam.2020.100922 - PubMed
-
- Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

