Characterization of a novel subfamily 1.4 lipase from Bacillus licheniformis IBRL-CHS2: Cloning and expression optimization
- PMID: 39689112
- PMCID: PMC11651597
- DOI: 10.1371/journal.pone.0314556
Characterization of a novel subfamily 1.4 lipase from Bacillus licheniformis IBRL-CHS2: Cloning and expression optimization
Abstract
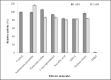
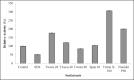
This study focuses on a novel lipase from Bacillus licheniformis IBRL-CHS2. The lipase gene was cloned into the pGEM-T Easy vector, and its sequences were registered in GenBank (KU984433 and AOT80658). It was identified as a member of the bacterial lipase subfamily 1.4. The pCold I vector and E. coli BL21 (DE3) host were utilized for expression, with the best results obtained by removing the enzyme's signal peptide. Optimal conditions were found to be 15°C for 24 h, using 0.2 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). The His-tagged lipase was purified 13-fold with a 68% recovery and a specific activity of 331.3 U/mg using affinity purification. The lipase demonstrated optimal activity at 35°C and pH 7. It remained stable after 24 h in 25% (v/v) organic solvents such as isooctane, n-hexane, dimethyl sulfoxide (DMSO), and methanol, which enhanced its activity. Chloroform and diethyl ether inhibited the lipase. The enzyme exhibited the highest affinity for p-nitrophenol laurate (C12:0) with a Km of 0.36 mM and a Vmax of 357 μmol min-1 mg-1. Among natural oils, it performed best with coconut oil and worst with olive oil. The lipase was stable in the presence of 1 mM and 5 mM Ca2⁺, K⁺, Na⁺, Mg2⁺, and Ba2⁺, but its activity decreased with Zn2⁺ and Al3⁺. Non-ionic surfactants like Triton X-100, Nonidet P40, Tween 20, and Tween 40 boosted activity, while Sodium Dodecyl Sulfate (SDS) inhibited it. This lipase's unique properties, particularly its stability in organic solvents, make it suitable for applications in organic synthesis and various industries.
Copyright: © 2024 Khazaal Kadhim Almansoori et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that we do not have any conflict of interest to the content of this article.
Figures



References
-
- Maleki A., Hassanzadeh-Afruzi F., Varzi Z., & Esmaeili M. S. (2020). Magnetic dextrin nanobiomaterial: An organic-inorganic hybrid catalyst for the synthesis of biologically active polyhydroquinoline derivatives by asymmetric Hantzsch reaction. Mater. Sci. Eng. C, 109, 110502. doi: 10.1016/j.msec.2019.110502 - DOI - PubMed
-
- Mehta A., Guleria S., Sharma R., & Gupta R. (2021). The lipases and their applications with emphasis on food industry. In Microbial Biotechnology in Food and Health (pp. 143–164). Elsevier. 10.1016/B978-0-12-819813-1.00006-2. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

