Genome alteration of Leishmania orientalis under Amphotericin B inhibiting conditions
- PMID: 39689148
- PMCID: PMC11687891
- DOI: 10.1371/journal.pntd.0012716
Genome alteration of Leishmania orientalis under Amphotericin B inhibiting conditions
Abstract
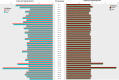
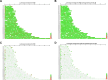
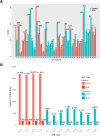
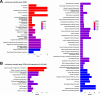
Amphotericin B (AmB) is a potent antifungal and antiparasitic medication that exerts its action by disrupting the cell membrane of the leishmanial parasite, leading to its death. Understanding the genetic alterations induced by Amphotericin B is crucial for gaining insights into drug resistance mechanisms and developing more effective treatments against Leishmania infections. As a new Leishmania species, the molecular response of Leishmania orientalis to anti-leishmanial drugs has not been fully explored. In this study, Leishmania orientalis strain PCM2 culture was subjected to AmB exposure at a concentration of 0.03 uM over 72 hours compared to the control. The genomic alteration and transcriptomic changes were investigated by utilising the whole genome and RNA sequencing methods, followed by the analysis of single nucleotide polymorphisms (SNPs), differential gene expression, and chromosomal copy number variations (CNVs) assessed using read depth coverage (RDC) values across the entire genome. The chromosomal CNV analysis showed no significant difference between L. orientalis from the control and AmB-treated groups. The distribution of SNPs displayed notable variability, with higher SNP incidence in the control group compared to the AmB-treated group. Gene ontology analysis unveiled functions of the SNPs -associated genes involved in transporter function, genetic precursor synthesis, and purine nucleotide metabolism. Notably, the impact of AmB treatment on the L. orientalis gene expression profiles exhibited diverse expressional alterations, particularly the downregulation of pivotal genes such as the tubulin alpha chain gene. The intricate interplay between SNPs and gene expression alterations might underscore the complex regulatory networks underlying the AmB resistance of L. orientalis strain PCM2.
Copyright: © 2024 Anuntasomboon et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Beverley SM. Gene amplification in Leishmania. Annual review of microbiology. 1991;45(1):417–44. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

