High-throughput specificity profiling of antibody libraries using ribosome display and microfluidics
- PMID: 39689695
- PMCID: PMC11704616
- DOI: 10.1016/j.crmeth.2024.100934
High-throughput specificity profiling of antibody libraries using ribosome display and microfluidics
Abstract
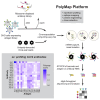
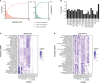
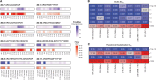
In this work, we developed PolyMap (polyclonal mapping), a high-throughput method for mapping protein-protein interactions. We demonstrated the mapping of thousands of antigen-antibody interactions between diverse antibody libraries isolated from convalescent and vaccinated COVID-19 donors and a set of clinically relevant SARS-CoV-2 spike variants. We identified over 150 antibodies with a variety of distinctive binding patterns toward the antigen variants and found a broader binding profile, including targeting of the Omicron variant, in the antibody repertoires of more recent donors. We then used these data to select mixtures of a small number of clones with complementary reactivity that together provide strong potency and broad neutralization. PolyMap is a generalizable platform that can be used for one-pot epitope mapping, immune repertoire profiling, and therapeutic design and, in the future, could be expanded to other families of interacting proteins.
Keywords: CP: Biotechnology; CP: Immunology; PolyMap; SARS-CoV-2 surface antigen; antibody-antigen interaction; cell surface protein; high-throughput screening; polyclonal antibody profiling; protein-protein interaction; ribosome display; single-cell analysis; viral neutralization.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests All authors are current or past employees of GigaGen, Inc. (a Grifols company). E.K.W., K.P.C., Y.W.L., A.S.A., and J.F.S. are inventors on a patent application related to the PolyMap platform.
Figures


References
-
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. - DOI - PMC - PubMed
-
- Li M., Beck E.J., Laeyendecker O., Eby Y., Tobian A.A.R., Caturegli P., Wouters C., Chiklis G.R., Block W., McKie R.O., et al. Convalescent plasma with a high level of virus-specific antibody effectively neutralizes SARS-CoV-2 variants of concern. Blood Adv. 2022;6:3678–3683. doi: 10.1182/bloodadvances.2022007410. - DOI - PMC - PubMed
-
- Wayham N.P., Niedecken A.R., Simons J.F., Chiang Y.Y., Medina-Cucurella A.V., Mizrahi R.A., Wagner E.K., Gras A., Segal I., Witte P., et al. A Potent Recombinant Polyclonal Antibody Therapeutic for Protection Against New Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern. J. Infect. Dis. 2023;228:555–563. doi: 10.1093/infdis/jiad102. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

