T-cell diversity and exclusion of blood-derived T-cells in the tumor microenvironment of classical Hodgkin Lymphoma
- PMID: 39690183
- PMCID: PMC11879864
- DOI: 10.1038/s41375-024-02490-6
T-cell diversity and exclusion of blood-derived T-cells in the tumor microenvironment of classical Hodgkin Lymphoma
Abstract
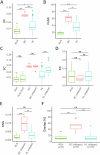
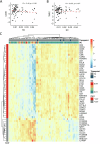
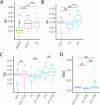
The Tumor Microenvironment (TME) in classical Hodgkin Lymphoma (HL) contains abundant immune cells and only few neoplastic Hodgkin and Reed-Sternberg cells (HRSC). We analyzed the T-cell receptor (TCR) repertoire to detect T-cell expansion in the TME and blood. In contrast to solid cancer tissue, T-cells in the TME of HL are highly polyclonal at first diagnosis and show only minor clonal expansion during anti-PD1 immune checkpoint blockade (ICB). At relapse and during ICB, pre-amplified T-cell populations increase in the TME of solid cancers but to a much lesser extent in HL. In contrast, T-cell populations in the peripheral blood of HL patients display higher clonality than healthy controls reaching clonality levels comparable to solid cancer. However, pre-amplified blood T-cells in HL patients show only minor additional clonal expansion during ICB. Moreover, blood-derived T-cells do not repopulate the TME of HL to the same extent as observed in solid cancers. Thus, the T-cell repertoire in the TME of HL appears unique by a relatively low clonal T-cell content and the exclusion of clonally expanded T-cells from the peripheral blood. Exclusion of clonally expanded tumor-specific T-cells from the TME may present a novel mechanism of immune evasion in HL.
© 2024. The Author(s).
Conflict of interest statement
COMPETING INTERESTS: PB reports grants from BMS during the conduct of the study. No other disclosures were reported. PJB is an advisor or consultant for Merck Sharp & Dohme, Need Inc., Stemline, and Takeda; holds stock options in Need Inc., has received honoraria from BeiGene, BMS/Celgene, Merck Sharp & Dohme, Need Inc., Stemline and Takeda and reports research funding from BeiGene (Inst), BMS (Inst), Merck Sharp & Dohme (Inst) and Takeda (Inst); all outside the submitted work. WK reports grants from Roche, Amgen, Takeda, inCyte, and Regeneron paid to his institution not related to the submitted work. No other potential conflicts of interest are reported. TH received honoraria from Amgen, Astra Zeneca, Daiichi Sankyo, Exact Sciences, Gilead, Lilly, MSD, Novartis, Roche, Serag Wiessner, and travel support from Gilead. Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. Approval has been obtained from the ethics advisory committee of the Medical Faculty of the University of Kiel (D464/17). Informed consent was obtained from all participants of the NIVAHL trial.
Figures


References
-
- Jachimowicz RD, Pieper L, Reinke S, Gontarewicz A, Plütschow A, Haverkamp H, et al. Analysis of the tumor microenvironment by whole-slide image analysis identifies low B cell content as a predictor of adverse outcome in advanced-stage classical Hodgkin lymphoma treated with BEACOPP. Haematologica. 2021;106:1684–92. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

