Proteogenomic characterization of highly enriched viable leukemic blasts in acute myeloid leukemia: A SWOG report
- PMID: 39691254
- PMCID: PMC11647701
- DOI: 10.1002/jha2.1041
Proteogenomic characterization of highly enriched viable leukemic blasts in acute myeloid leukemia: A SWOG report
Abstract
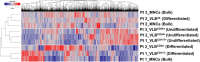
Introduction: Acute myeloid leukemia (AML) remains one of the deadliest hematopoietic malignancies. A better understanding of the molecular biology governing AML may lead to improved risk stratification and facilitate the development of novel therapies. Proteins are responsible for much of the biology of cells. Several studies have examined the global proteome in bulk mononuclear cells (MNCs) from AML specimens, which are comprised a heterogenous population of cells at various stages of differentiation.
Methods: Given the potential impact of the nonleukemic cells on protein expression profiles, we applied an integrative proteogenomic approach utilizing next-generation sequencing and mass spectrometry-based proteomics to identify novel protein biomarkers in unsorted MNCs and viable leukemic blasts (VLBs) isolated from blood and bone marrow specimens obtained at the time of AML diagnosis.
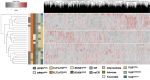
Results: We identified significant differences in protein expression between VLBs and MNCs. Subsequent studies (N = 27) focused on proteomic profiling of VLBs that identified novel candidate biomarkers associated with mutational genotypes and clinical outcome, some of which were recapitulated in an independent cohort of patients. Using mass spectrometry, we also detected mutated protein products, some of which were predicted via in silico analyses to be potential neoantigens amenable to adoptive immunotherapy. As previously described, analyses comparing transcript and protein expression showed an overall modest correlation between mRNA and protein dataset, but enriching for genes associated with mutations significantly improved the protein-RNA correlation.
Conclusion: Together, the results provide insight into the biology of VLBs and demonstrate the gains derived from examining the proteome in addition to genome and transcriptome.
Keywords: AML; hematological malignancy; neoantigens; proteogenomics; transcriptomics.
© 2024 The Author(s). eJHaem published by British Society for Haematology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Verification of prognostic expression biomarkers is improved by examining enriched leukemic blasts rather than mononuclear cells from acute myeloid leukemia patients.Biomark Res. 2023 Mar 16;11(1):31. doi: 10.1186/s40364-023-00461-0. Biomark Res. 2023. PMID: 36927800 Free PMC article.
-
Impact of Specimen Heterogeneity on Biomarkers in Repository Samples from Patients with Acute Myeloid Leukemia: A SWOG Report.Biopreserv Biobank. 2018 Feb;16(1):42-52. doi: 10.1089/bio.2017.0079. Epub 2017 Nov 27. Biopreserv Biobank. 2018. PMID: 29172682 Free PMC article.
-
Proteogenomic profiling of acute myeloid leukemia to identify therapeutic targets.Expert Rev Proteomics. 2024 Nov 22:1-14. doi: 10.1080/14789450.2024.2431272. Online ahead of print. Expert Rev Proteomics. 2024. PMID: 39576246
-
Proteogenomics approaches for studying cancer biology and their potential in the identification of acute myeloid leukemia biomarkers.Expert Rev Proteomics. 2017 Aug;14(8):649-663. doi: 10.1080/14789450.2017.1352474. Epub 2017 Jul 27. Expert Rev Proteomics. 2017. PMID: 28693350 Review.
-
Neoantigens in Hematological Malignancies-Ultimate Targets for Immunotherapy?Front Immunol. 2019 Dec 20;10:3004. doi: 10.3389/fimmu.2019.03004. eCollection 2019. Front Immunol. 2019. PMID: 31921218 Free PMC article. Review.
References
-
- Luczak M, Kazmierczak M, Handschuh L, Lewandowski K, Komarnicki M, Figlerowicz M. Comparative proteome analysis of acute myeloid leukemia with and without maturation. J Proteomics. 2012;75(18):5734–5748. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
