Recognition and Localization of Maize Leaf and Stalk Trajectories in RGB Images Based on Point-Line Net
- PMID: 39691278
- PMCID: PMC11651416
- DOI: 10.34133/plantphenomics.0199
Recognition and Localization of Maize Leaf and Stalk Trajectories in RGB Images Based on Point-Line Net
Abstract
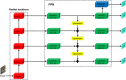
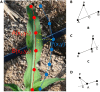
Plant phenotype detection plays a crucial role in understanding and studying plant biology, agriculture, and ecology. It involves the quantification and analysis of various physical traits and characteristics of plants, such as plant height, leaf shape, angle, number, and growth trajectory. By accurately detecting and measuring these phenotypic traits, researchers can gain insights into plant growth, development, stress tolerance, and the influence of environmental factors, which has important implications for crop breeding. Among these phenotypic characteristics, the number of leaves and growth trajectory of the plant are most accessible. Nonetheless, obtaining these phenotypes is labor intensive and financially demanding. With the rapid development of computer vision technology and artificial intelligence, using maize field images to fully analyze plant-related information can greatly eliminate repetitive labor and enhance the efficiency of plant breeding. However, it is still difficult to apply deep learning methods in field environments to determine the number and growth trajectory of leaves and stalks due to the complex backgrounds and serious occlusion problems of crops in field environments. To preliminarily explore the application of deep learning technology to the acquisition of the number of leaves and stalks and the tracking of growth trajectories in field agriculture, in this study, we developed a deep learning method called Point-Line Net, which is based on the Mask R-CNN framework, to automatically recognize maize field RGB images and determine the number and growth trajectory of leaves and stalks. The experimental results demonstrate that the object detection accuracy (mAP50) of our Point-Line Net can reach 81.5%. Moreover, to describe the position and growth of leaves and stalks, we introduced a new lightweight "keypoint" detection branch that achieved a magnitude of 33.5 using our custom distance verification index. Overall, these findings provide valuable insights for future field plant phenotype detection, particularly for datasets with dot and line annotations.
Copyright © 2024 Bingwen Liu et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures







References
-
- Mangelsdorf PC. The origin and evolution of maize. Adv Genet. 1947;1:161–207. - PubMed
-
- Brown LR. The world outlook for conventional agriculture. More emphasis is needed on farm price policy and plant research if future world food needs are to be met. Science. 1967;158(3801):604–611. - PubMed
-
- Stokstad E. High hopes for short corn. Science. 2023;382(6669):364–367. - PubMed
-
- Nabizadeh E, Banifazel M, Taherifard E. The effects of plant growth promoting on some of traits in maize (cv. SC 704) under drought stress condition. European. J Exp Biol. 2012;2(4):875–881.
LinkOut - more resources
Full Text Sources

