Quantitative proteomic analysis of residual host cell protein retention across adeno-associated virus affinity chromatography
- PMID: 39691383
- PMCID: PMC11650319
- DOI: 10.1016/j.omtm.2024.101383
Quantitative proteomic analysis of residual host cell protein retention across adeno-associated virus affinity chromatography
Abstract
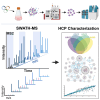
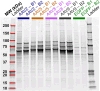
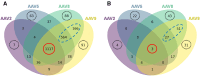
To better understand host cell protein (HCP) retention in adeno-associated virus (AAV) downstream processes, sequential window acquisition of all theoretical fragment ion mass spectra (SWATH-MS) was used to quantitatively profile residual HCPs for four AAV serotypes (AAV2, -5, -8, and -9) produced with HEK293 cells and purified using POROS CaptureSelect AAVX affinity chromatography. A broad range of residual HCPs were detected in affinity eluates after purification (N total = 2,746), and HCP profiles showed universally present species (N universal = 1,117) and species unique to one or more AAV serotype. SWATH-MS revealed that HCP persistence was dominated by high-abundance conserved species (HACS), which appeared across all serotype conditions studied. Due to the notable contribution of these species to overall residual HCP levels, physical and functional characteristics of HACS were examined to determine trends that coincide with persistence. Subnetwork interaction mapping and Gene Ontology function enrichment analysis revealed extensive physical interactions between these proteins and significant enrichment for biological processes, molecular functions, and reactome pathways related to protein folding, nucleic acid binding, and cellular stress. The abundant and conserved nature of these HCPs and their functions offers a new perspective for mechanistic evaluations of impurity retention for AAV downstream processes.
Keywords: AAV; AAVX affinity chromatography; HCP; LC-MS/MS; adeno-associated virus; host cell protein; liquid chromatography-tandem mass spectrometry; proteomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Clinicaltrials.gov (accessed November 8, 2023).
-
- Dobrowsky T., Gianni D., Pieracci J., Suh J. AAV manufacturing for clinical use: Insights on current challenges from the upstream process. Current Opinion in Biomedical Engineering. 2021;20 doi: 10.1016/j.cobme.2021.100353. - DOI
-
- Zhao H., Lee K.J., Daris M., Lin Y., Wolfe T., Sheng J., Plewa C., Wang S., Meisen W.H. Creation of a High-Yield AAV Vector Production Platform in Suspension Cells Using a Design-of-Experiment Approach. Mol. Ther. Methods Clin. Dev. 2020;18:312–320. doi: 10.1016/j.omtm.2020.06.004. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

