The interplay between epitranscriptomic RNA modifications and neurodegenerative disorders: Mechanistic insights and potential therapeutic strategies
- PMID: 39691424
- PMCID: PMC11649393
- DOI: 10.1002/ibra.12183
The interplay between epitranscriptomic RNA modifications and neurodegenerative disorders: Mechanistic insights and potential therapeutic strategies
Abstract
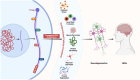
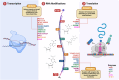
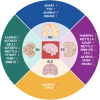
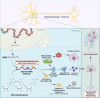
Neurodegenerative disorders encompass a group of age-related conditions characterized by the gradual decline in both the structure and functionality of the central nervous system (CNS). RNA modifications, arising from the epitranscriptome or RNA-modifying protein mutations, have recently been observed to contribute significantly to neurodegenerative disorders. Specific modifications like N6-methyladenine (m6A), N1-methyladenine (m1A), 5-methylcytosine (m5C), pseudouridine and adenosine-to-inosine (A-to-I) play key roles, with their regulators serving as crucial therapeutic targets. These epitranscriptomic changes intricately control gene expression, influencing cellular functions and contributing to disease pathology. Dysregulation of RNA metabolism, affecting mRNA processing and noncoding RNA biogenesis, is a central factor in these diseases. This review underscores the complex relationship between RNA modifications and neurodegenerative disorders, emphasizing the influence of RNA modification and the epitranscriptome, exploring the function of RNA modification enzymes in neurodegenerative processes, investigating the functional consequences of RNA modifications within neurodegenerative pathways, and evaluating the potential therapeutic advancements derived from assessing the epitranscriptome.
Keywords: Alzheimer's disease; Parkinson's disease; RNA modifications; epitranscriptomics; neurodegeneration.
© 2024 The Author(s). Ibrain published by Affiliated Hospital of Zunyi Medical University (AHZMU) and Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no competing interests associated with the publication of this review article.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
