Exploring the Microbial Landscape of Bone and Joint Infections: An Analysis Using 16S rRNA Metagenome Sequencing
- PMID: 39691489
- PMCID: PMC11651062
- DOI: 10.2147/IDR.S482931
Exploring the Microbial Landscape of Bone and Joint Infections: An Analysis Using 16S rRNA Metagenome Sequencing
Abstract
Background: Bone and joint infections (BJIs) are challenging to diagnose. This study evaluated the utility of 16S rRNA gene sequencing in diagnosing BJIs, comparing it with conventional bacterial culture to explore microbial diversity in orthopedic infections.
Methods: Thirty patients with BJIs were enrolled from January 2019 to September 2020 at a single orthopedic center. Diagnoses were based on the Musculoskeletal Infection Society standards. DNA extraction, 16S rRNA sequencing, and microbial composition analysis were performed. Conventional bacterial culture results were compared with metagenomics detection, and associations with blood routine and biochemical test factors were analyzed.
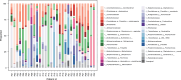
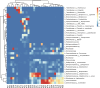
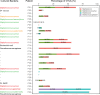
Results: The study enrolled 30 patients with BJIs. Traditional bacterial culture successfully identified pathogens in 60% (18/30) of cases, predominantly Staphylococcus aureus. In contrast, 16S rRNA metagenomics sequencing revealed distinct microorganisms in all cases, it unveiled a diverse microbial landscape. The correlation between bacterial culture and metagenomics detection showcased both concordance and discrepancies. Consistency of detection between the two methods showed that metagenomics detection detected the same genus or species in 14 (87.5%) of the 16 samples identified as species by bacterial culture. In nearly half of the patients with negative cultures, pathogenic microorganisms were detected, highlighting the capability of 16S rRNA sequencing to identify microorganisms, even in samples with negative or unidentified culture results. Moreover, no significant correlation was observed between bacterial culture, metagenomics detection and the factors of blood routine and biochemical test.
Conclusion: This study deepens our understanding of the microbial complexity in BJIs. While traditional culture methods are cost-effective and practical, 16S rRNA gene sequencing proves valuable for complementary microbial analysis, particularly when traditional methods fail or rapid identification is critical. This emerging diagnostic approach can enhance the accuracy and speed of pathogen identification, enabling more effective interventions in the management of BJIs.
Keywords: 16S rRNA gene sequencing; bone and joint infections; clinical management; diagnostic tool; microbial complexity.
© 2024 Maimaiti and Liu.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources

