Resistome in the indoor dust samples from workplaces and households: a pilot study
- PMID: 39691696
- PMCID: PMC11649746
- DOI: 10.3389/fcimb.2024.1484100
Resistome in the indoor dust samples from workplaces and households: a pilot study
Abstract
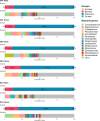
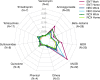
The antibiotic resistance genes (ARGs) limit the susceptibility of bacteria to antimicrobials, representing a problem of high importance. Current research on the presence of ARGs in microorganisms focuses mainly on humans, livestock, hospitals, or wastewater. However, the spectrum of ARGs in the dust resistome in workplaces and households has gone relatively unexplored. This pilot study aimed to analyze resistome in indoor dust samples from participants' workplaces (a pediatric hospital, a maternity hospital, and a research center) and households and compare two different approaches to the ARGs analysis; high-throughput quantitative PCR (HT-qPCR) and whole metagenome shotgun sequencing (WMGS). In total, 143 ARGs were detected using HT-qPCR, with ARGs associated with the macrolides, lincosamides, and streptogramin B (MLSB) phenotype being the most abundant, followed by MDR (multi-drug resistance) genes, and genes conferring resistance to aminoglycosides. A higher overall relative quantity of ARGs was observed in indoor dust samples from workplaces than from households, with the pediatric hospital being associated with the highest relative quantity of ARGs. WMGS analysis revealed 36 ARGs, of which five were detected by both HT-qPCR and WMGS techniques. Accordingly, the efficacy of the WMGS approach to detect ARGs was lower than that of HT-qPCR. In summary, our pilot data revealed that indoor dust in buildings where people spend most of their time (workplaces, households) can be a significant source of antimicrobial-resistant microorganisms, which may potentially pose a health risk to both humans and animals.
Keywords: antibiotic resistance gene; antimicrobial resistance; hospital; indoor environment; microbiome.
Copyright © 2024 Klvanova, Videnska, Barton, Bohm, Splichalova, Koksova, Urik, Lanickova, Prokes, Budinska, Klanova and Borilova Linhartova.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Department-specific patterns of bacterial communities and antibiotic resistance in hospital indoor environments.Appl Microbiol Biotechnol. 2024 Oct 16;108(1):487. doi: 10.1007/s00253-024-13326-9. Appl Microbiol Biotechnol. 2024. PMID: 39412549 Free PMC article.
-
Metagenomic airborne resistome from urban hot spots through the One Health lens.Environ Microbiol Rep. 2024 Jun;16(3):e13306. doi: 10.1111/1758-2229.13306. Environ Microbiol Rep. 2024. PMID: 38923122 Free PMC article.
-
Antimicrobial resistance genes in the oral microbiome.Evid Based Dent. 2025 Mar;26(1):42-43. doi: 10.1038/s41432-025-01120-z. Epub 2025 Mar 6. Evid Based Dent. 2025. PMID: 40050500 Free PMC article.
-
Profiles of Microbial Community and Antibiotic Resistome in Wild Tick Species.mSystems. 2022 Aug 30;7(4):e0003722. doi: 10.1128/msystems.00037-22. Epub 2022 Aug 1. mSystems. 2022. PMID: 35913190 Free PMC article. Review.
-
Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review.Molecules. 2019 Jan 3;24(1):163. doi: 10.3390/molecules24010163. Molecules. 2019. PMID: 30609875 Free PMC article. Review.
References
-
- Abdelrahman D. N., Taha A. A., Dafaallah M. M., Mohammed A. A., El Hussein A. R. M., Hashim A. I., et al. . (2020). [amp]]beta;-lactamases (bla TEM, bla SHV, bla CTXM-1, bla VEB, bla OXA-1) and class C β-lactamases gene frequency in Pseudomonas aeruginosa isolated from various clinical specimens in Khartoum State, Sudan: a cross sectional study. F1000Research 9, 774. doi: 10.12688/f1000research.24818.3 - DOI - PMC - PubMed
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed June 29, 2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

